The SPI-3 pathogenicity island of Salmonella enterica
- PMID: 9922266
- PMCID: PMC93469
- DOI: 10.1128/JB.181.3.998-1004.1999
The SPI-3 pathogenicity island of Salmonella enterica
Abstract
Pathogenicity islands are chromosomal clusters of pathogen-specific virulence genes often found at tRNA loci. We have determined the molecular genetic structure of SPI-3, a 17-kb pathogenicity island located at the selC tRNA locus of Salmonella enterica serovar Typhimurium. The G+C content of SPI-3 (47.5%) differs from that of the Salmonella genome (52%), consistent with the notion that these sequences have been horizontally acquired. SPI-3 harbors 10 open reading frames organized in six transcriptional units, which include the previously described mgtCB operon encoding the macrophage survival protein MgtC and the Mg2+ transporter MgtB. Among the newly identified open reading frames, one exhibits sequence similarity to the ToxR regulatory protein of Vibrio cholerae and one is similar to the AIDA-I adhesin of enteropathogenic Escherichia coli. The distribution of SPI-3 sequences varies among the salmonellae: the right end of the island, which harbors the virulence gene mgtC, is present in all eight subspecies of Salmonella; however, a four-gene cluster at the center of SPI-3 is found in only some of the subspecies and is bracketed by remnants of insertion sequences, suggesting a multistep process in the evolution of SPI-3 sequences.
Figures


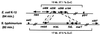
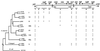
References
-
- Bajaj V, Hwang C, Lee C A. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol. 1995;18:715–727. - PubMed
-
- Benz I, Schmidt M A. AIDA-I, the adhesin involved in diffuse adherence of the diarrhoeagenic Escherichia coli strain 2787 (O126:H27), is synthesized via a precursor molecule. Mol Microbiol. 1992;6:1539–1546. - PubMed
-
- Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

