IS6110-mediated deletions of wild-type chromosomes of Mycobacterium tuberculosis
- PMID: 9922268
- PMCID: PMC93471
- DOI: 10.1128/JB.181.3.1014-1020.1999
IS6110-mediated deletions of wild-type chromosomes of Mycobacterium tuberculosis
Abstract
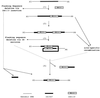
The ipl locus is a site for the preferential insertion of IS6110 and has been identified as an insertion sequence, IS1547, in its own right. Various deletions around the ipl locus of clinical isolates of Mycobacterium tuberculosis were identified, and these deletions ranged in length from several hundred base pairs up to several kilobase pairs. The most obvious feature shared by these deletions was the presence of an IS6110 copy at the deletion sites, which suggested two possible mechanisms for their occurrence, IS6110 transposition and homologous recombination. To clarify the mechanism, an investigation was conducted; the results suggest that although deletion transpositionally mediated by IS6110 was a possibility, homologous recombination was a more likely one. The implications of such chromosomal rearrangements for the evolution of M. tuberculosis, for IS6110-mediated mutagenesis, and for the development of genetic tools are discussed. The deletion of genomic DNA in isolates of M. tuberculosis has previously been noted at only a few sites. This study examined the deletional loss of genetic material at a new site and suggests that such losses may occur elsewhere too and may be more prevalent than was previously thought. Distinct from the study of laboratory-induced mutations, the detailed analysis of clinical isolates, in combination with knowledge of their evolutionary relationships to each other, gives us the opportunity to study mutational diversity in isolates that have survived in the human host and therefore offers a different perspective on the importance of particular genetic markers in pathogenesis.
Figures





References
-
- Bernardi A, Bernardi F. Site-specific deletions in the recombinant plasmid pSC101 containing the redB-ori region of phage lambda. Gene. 1981;13:103–109. - PubMed
-
- Caspers P, Dalrymple B, Iida S, Arber W. IS30, a new insertion sequence of Escherichia coli K12. Mol Gen Genet. 1984;196:68–73. - PubMed
-
- Collins D M, Stephens D M. Identification of an insertion sequence, IS1081, in Mycobacterium bovis. FEMS Microbiol Lett. 1991;67:11–15. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

