Genetic methods for assessing antimicrobial resistance
- PMID: 9925507
- PMCID: PMC89052
- DOI: 10.1128/AAC.43.2.199
Genetic methods for assessing antimicrobial resistance
Figures
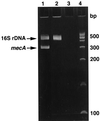


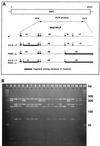

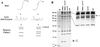



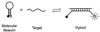

References
-
- Anonymous. Methods of creating DNA molecules. In: Watson J D, Gilman M, Witkowski J, Moller Z, editors. Recombinant DNA. New York, N.Y: Scientific American Books; 1992. pp. 63–75.
-
- Arlet G, Brami G, Décrè D, Flippo A, Gaillot O, Lagrange P H, Philippon A. Molecular characterization by PCR-restriction fragmented length polymorphism of TEM β-lactamases. FEMS Microbiol Let. 1995;134:203–208. - PubMed
-
- Ausubel F M, Albright L M. DNA sequencing. In: Ausubel F, Brent R, Kingston R, Moore D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology, suppl. 23. Boston, Mass: John Wiley and Sons; 1998. pp. 7.0.1–7.7.7.23.
-
- Bignardi G E, Woodford N, Chapman A, Johnson A P, Speller D C E. Detection of the mecA gene and phenotypic detection of resistance in Staphylococcus aureus isolates with borderline or low-level methicillin resistance. J Antimicrob Chemother. 1996;37:53–63. - PubMed
-
- Brow M A, Oldenburg M C, Lyamichev V, Heisler L M, Lyamichera N, Hall J G, Eagen N J, Olive D M, Smith L M, Fors L, Dahlberg J E. Differentiation of bacterial 16S rRNA genes and intergenic regions and Mycobacterium tuberculosis katG genes by structure-specific endonuclease cleavage. J Clin Microbiol. 1996;34:2139–3132. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

