glmM operon and methicillin-resistant glmM suppressor mutants in Staphylococcus aureus
- PMID: 9925512
- PMCID: PMC89057
- DOI: 10.1128/AAC.43.2.240
glmM operon and methicillin-resistant glmM suppressor mutants in Staphylococcus aureus
Abstract
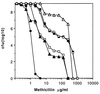
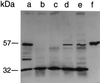
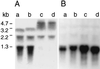
The Staphylococcus aureus phosphoglucosamine mutase gene glmM was shown to be the last gene of a three-cistron operon, orf1-orf2-glmM. One transcriptional start was identified upstream of orf1, and a second start producing a monocistronic transcript was identified upstream of glmM. Disruption of glmM abolished GlmM production, decreased methicillin resistance, and resulted in teicoplanin hypersusceptibility without affecting the production of the endogenous penicillin-binding proteins and PBP 2'. Complementation of the glmM mutation by the complete glmM operon restored both methicillin resistance and normal teicoplanin susceptibility. In contrast, a highly methicillin-resistant suppressor mutant obtained by selection for growth in the presence of methicillin remained GlmM deficient and teicoplanin hypersusceptible. The suppressor mutation was not linked to the glmM operon but was correlated with decreased autolysis and increased production of a 49-kDa protein, suggesting that there is an alternative pathway for glucosamine-1-phosphate synthesis in S. aureus.
Figures



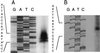
References
-
- Berger-Bächi B, Barberis-Maino L, Strässle A, Kayser F H. FemA, a host-mediated factor essential for methicillin resistance in Staphylococcus aureus: molecular cloning and characterization. Mol Gen Genet. 1989;219:263–269. - PubMed
-
- Berger-Bächi B, Kohler M L. A novel site on the chromosome of Staphylococcus aureus influencing the level of methicillin resistance: genetic mapping. FEMS Microbiol Lett. 1983;20:305–309.
-
- Berger-Bächi B, Strässle A, Kayser F H. Characterization of an isogenic set of methicillin-resistant and -susceptible mutants of Staphylococcus aureus. Eur J Clin Microbiol. 1986;5:697–701. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

