Genetic variation and susceptibilities to protease inhibitors among subtype B and F isolates in Brazil
- PMID: 9925514
- PMCID: PMC89059
- DOI: 10.1128/AAC.43.2.253
Genetic variation and susceptibilities to protease inhibitors among subtype B and F isolates in Brazil
Abstract
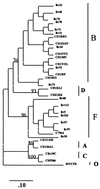
The genetic variation of the human immunodeficiency virus type 1 (HIV-1) protease gene (prt) permits the classification of HIV-1 strains into five distinct protease subtypes, which follow the gag subtyping patterns. The susceptibilities of non-B-subtype strains to protease inhibitors (PIs) and other antiretroviral drugs remain largely unknown. Subtype F is the main non-B strain contributing to the Brazilian epidemic, accounting for 15 to 20% of these infections. In this work, we report the findings on 81 isolates from PI-naive Brazilian patients collected between 1993 and 1997. In addition, the relevant PI resistance mutations and their phenotypes were determined in vitro for 15 of these patients (B = 9 and F = 6). Among these, the subtype F samples evidenced high sensitivities in vitro to ritonavir and indinavir, with MICs at which 50 and 90% of the isolates are inhibited similar to those of both the Brazilian and the U.S. subtype B isolates. Analysis of the 81 Brazilian prt sequences demonstrated that the subtype F consensus sequence differs from the U.S. and Brazilian subtype B consensus in eight positions (I15V, E35D, M36I, R41K, R57K, Q61N, L63P, and L89M). The frequency of critical PI resistance substitutions (amino acid changes D30N, V82A/F/T, I84V, N88D, and L90M) among Brazilian isolates is very low (mean, 2.5%), and the associated secondary substitutions (amino acid positions 10L, 20K, 36M, 46M, 48G, 54I, 63P, 71A, and 77A) are infrequent. These observations document the relative rarity of resistance to PIs in the treatment of patients infected with HIV-1 subtype F in South America.
Figures

References
-
- Brodine S K, Mascola J R, Weiss P J, Ito S I, Porter K R, Artenstein A W, Garland F C, McCutchan F E, Burke D S. Detection of diverse HIV-1 genetic subtypes in the USA. Lancet. 1995;346:1198–1199. - PubMed
-
- Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1996;271:670–671. - PubMed
-
- Condra J H, Holder D J, Schleif W A, Blahy O M, Danovich R M, Gabryelski L J, Graham D J, Laird D, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Yang T, Chodakewitz J A, Deutsch P J, Leavitt R Y, Massari F E, Mellors J W, Squires K E, Steigbigel R T, Teppler H, Emini E A. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J Virol. 1996;70:8270–8276. - PMC - PubMed
-
- da Costa S M, Schechter M, Shindo N, Vicente A C, Oliveira E F, Pinto M E, Tanuri A. Sequence and phylogenetic analysis of glycoprotein 120 of an HIV type 1 variant (GWGR) prevalent in Brazil. AIDS Res Hum Retroviruses. 1995;11:1143–1145. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

