Monitoring methanotrophic bacteria in hybrid anaerobic-aerobic reactors with PCR and a catabolic gene probe
- PMID: 9925557
- PMCID: PMC91036
- DOI: 10.1128/AEM.65.2.381-388.1999
Monitoring methanotrophic bacteria in hybrid anaerobic-aerobic reactors with PCR and a catabolic gene probe
Abstract
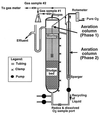
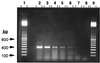
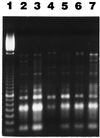
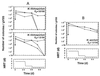
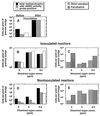
We attempted to mimic in small upflow anaerobic sludge bed (UASB) bioreactors the metabolic association found in nature between methanogens and methanotrophs. UASB bioreactors were inoculated with pure cultures of methanotrophs, and the bioreactors were operated by using continuous low-level oxygenation in order to favor growth and/or survival of methanotrophs. Unlike the reactors in other similar studies, the hybrid anaerobic-aerobic bioreactors which we used were operated synchronously, not sequentially. Here, emphasis was placed on monitoring various methanotrophic populations by using classical methods and also a PCR amplification assay based on the mmoX gene fragment of the soluble methane monooxygenase (sMMO). The following results were obtained: (i) under the conditions used, Methylosinus sporium appeared to survive better than Methylosinus trichosporium; (ii) the PCR method which we used could detect as few as about 2,000 sMMO gene-containing methanotrophs per g (wet weight) of granular sludge; (iii) inoculation of the bioreactors with pure cultures of methanotrophs contributed greatly to increases in the sMMO-containing population (although the sMMO-containing population decreased gradually with time, at the end of an experiment it was always at least 2 logs larger than the initial population before inoculation); (iv) in general, there was a good correlation between populations with the sMMO gene and populations that exhibited sMMO activity; and (v) inoculation with sMMO-positive cultures helped increase significantly the proportion of sMMO-positive methanotrophs in reactors, even after several weeks of operation under various regimes. At some point, anaerobic-aerobic bioreactors like those described here might be used for biodegradation of various chlorinated pollutants.
Figures
References
-
- American Public Health Association. Standard methods for the examination of water and wastewater. 16th ed. Washington, D.C: American Public Health Association; 1985.
-
- Bowman J P, Sayler G S. Optimization and maintenance of soluble methane monooxygenase activity in Methylosinus trichosporium OB3b. Biodegradation. 1994;5:1–11. - PubMed
-
- Brusseau G A, Tsien H C, Hanson R S, Wackett L P. Optimization of trichloroethylene oxidation by methanotrophs and the use of a colorimetric assay to detect soluble monooxygenase activity. Biodegradation. 1990;1:19–29. - PubMed
-
- Buchholz L A, Klump J V, Collins M L P, Brantner C A, Remsen C C. Activity of methanotrophic bacteria in Green Bay sediments. FEMS Microbiol Ecol. 1995;16:1–8.
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

