Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast
- PMID: 9925642
- PMCID: PMC316392
- DOI: 10.1101/gad.13.2.176
Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast
Abstract
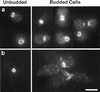
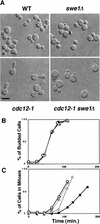
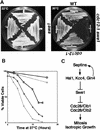
The mechanisms that couple cell cycle progression with the organization of the peripheral cytoskeleton are poorly understood. In Saccharomyces cerevisiae, the Swe1 protein has been shown previously to phosphorylate and inactivate the cyclin-dependent kinase, Cdc28, thereby delaying the onset of mitosis. The nim1-related protein kinase, Hsl1, induces entry into mitosis by negatively regulating Swe1. We have found that Hsl1 physically associates with the septin cytoskeleton in vivo and that Hsl1 kinase activity depends on proper septin function. Genetic analysis indicates that two additional Hsl1-related kinases, Kcc4 and Gin4, act redundantly with Hsl1 to regulate Swe1. Kcc4, like Hsl1 and Gin4, was found to localize to the bud neck in a septin-dependent fashion. Interestingly, hsl1 kcc4 gin4 triple mutants develop a cellular morphology extremely similar to that of septin mutants. Consistent with the idea that Hsl1, Kcc4, and Gin4 link entry into mitosis to proper septin organization, we find that septin mutants incubated at the restrictive temperature trigger a Swe1-dependent mitotic delay that is necessary to maintain cell viability. These results reveal for the first time how cells monitor the organization of their cytoskeleton and demonstrate the existence of a cell cycle checkpoint that responds to defects in the peripheral cytoskeleton. Moreover, Hsl1, Kcc4, and Gin4 have homologs in higher eukaryotes, suggesting that the regulation of Swe1/Wee1 by this class of kinases is highly conserved.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
