Transcriptional repression by the Caenorhabditis elegans germ-line protein PIE-1
- PMID: 9925644
- PMCID: PMC316391
- DOI: 10.1101/gad.13.2.202
Transcriptional repression by the Caenorhabditis elegans germ-line protein PIE-1
Abstract
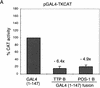
In the early Caenorhabditis elegans embryo, maternally expressed PIE-1 protein is required in germ-line blastomeres to inhibit somatic differentiation, maintain an absence of mRNA transcription, and block phosphorylation of the RNA polymerase II large subunit (Pol II) carboxy-terminal domain (CTD). We have determined that PIE-1 can function as a transcriptional repressor in cell culture assays. By fusing PIE-1 sequences to the yeast GAL4 DNA-binding domain, we have identified a PIE-1 repression domain that appears to inhibit the transcriptional machinery directly. A sequence element that is required for this repressor activity is similar to the Pol II CTD heptapeptide repeat, suggesting that the PIE-1 repression domain might target a protein complex that can bind the CTD. An alteration of this sequence element that blocks repression also impairs the ability of a transgene to rescue a pie-1 mutation, suggesting that this repressor activity may be important for PIE-1 function in vivo.
Figures









References
-
- Andersen B, Weinberg WC, Rennekampff O, McEvilly RJ, Bermingham JR, Jr, Hooshmand F, Vasilyev V, Hansbrough JF, Pittelkow MR, Yuspa SH, Rosenfeld MG. Functions of the POU domain genes Skn-1a/i and Tst-1/Oct-6/SCIP in epidermal differentiation. Genes & Dev. 1997;11:1873–1884. - PubMed
-
- Barabino SMLX, Hübner W, Jenny A, Minivielle-Sebastia L, Keller W. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes & Dev. 1997;11:1703–1716. - PubMed
-
- Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science. 1998;281:1001–1005. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
