Single-molecule fluorescence spectroscopy of enzyme conformational dynamics and cleavage mechanism
- PMID: 9927664
- PMCID: PMC15321
- DOI: 10.1073/pnas.96.3.893
Single-molecule fluorescence spectroscopy of enzyme conformational dynamics and cleavage mechanism
Abstract
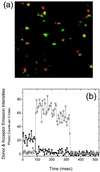
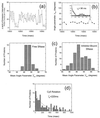
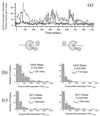
Fluorescence resonance energy transfer and fluorescence polarization anisotropy are used to investigate single molecules of the enzyme staphylococcal nuclease. Intramolecular fluorescence resonance energy transfer and fluorescence polarization anisotropy measurements of fluorescently labeled staphylococcal nuclease molecules reveal distinct patterns of fluctuations that may be attributed to protein conformational dynamics on the millisecond time scale. Intermolecular fluorescence resonance energy transfer measurements provide information about the dynamic interactions of staphylococcal nuclease with single substrate molecules. The experimental methods demonstrated here should prove generally useful in studies of protein folding and enzyme catalysis at single-molecule resolution.
Figures

References
-
- Funatsu T, Harada Y, Tokunaga M, Saito K, Yanagida T. Nature (London) 1995;374:555–559. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

