Detection of transient in vivo interactions between substrate and transporter during protein translocation into the endoplasmic reticulum
- PMID: 9950680
- PMCID: PMC25172
- DOI: 10.1091/mbc.10.2.329
Detection of transient in vivo interactions between substrate and transporter during protein translocation into the endoplasmic reticulum
Abstract
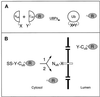
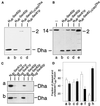
The split-ubiquitin technique was used to detect transient protein interactions in living cells. Nub, the N-terminal half of ubiquitin (Ub), was fused to Sec62p, a component of the protein translocation machinery in the endoplasmic reticulum of Saccharomyces cerevisiae. Cub, the C-terminal half of Ub, was fused to the C terminus of a signal sequence. The reconstitution of a quasi-native Ub structure from the two halves of Ub, and the resulting cleavage by Ub-specific proteases at the C terminus of Cub, serve as a gauge of proximity between the two test proteins linked to Nub and Cub. Using this assay, we show that Sec62p is spatially close to the signal sequence of the prepro-alpha-factor in vivo. This proximity is confined to the nascent polypeptide chain immediately following the signal sequence. In addition, the extent of proximity depends on the nature of the signal sequence. Cub fusions that bore the signal sequence of invertase resulted in a much lower Ub reconstitution with Nub-Sec62p than otherwise identical test proteins bearing the signal sequence of prepro-alpha-factor. An inactive derivative of Sec62p failed to interact with signal sequences in this assay. These in vivo findings are consistent with Sec62p being part of a signal sequence-binding complex.
Figures





References
-
- Beckmann R, Bubeck D, Grassucci R, Penczek P, Verschoor A, Blobel G, Frank J. Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex. Science. 1997;278:2123–2126. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

