Structure, properties, and tissue localization of apoplastic alpha-glucosidase in crucifers
- PMID: 9952433
- PMCID: PMC32114
- DOI: 10.1104/pp.119.2.385
Structure, properties, and tissue localization of apoplastic alpha-glucosidase in crucifers
Abstract
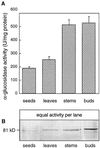
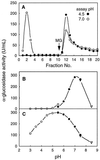
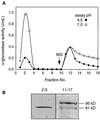
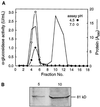
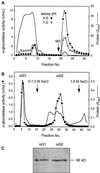
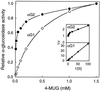
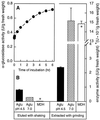
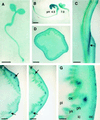
Apoplastic alpha-glucosidases occur widely in plants but their function is unknown because appropriate substrates in the apoplast have not been identified. Arabidopsis contains at least three alpha-glucosidase genes; Aglu-1 and Aglu-3 are sequenced and Aglu-2 is known from six expressed sequence tags. Antibodies raised to a portion of Aglu-1 expressed in Escherichia coli recognize two proteins of 96 and 81 kD, respectively, in vegetative tissues of Arabidopsis, broccoli (Brassica oleracea L.), and mustard (Brassica napus L.). The acidic alpha-glucosidase activity from broccoli flower buds was purified using concanavalin A and ion-exchange chromatography. Two active fractions were resolved and both contained a 96-kD immunoreactive polypeptide. The N-terminal sequence from the 96-kD broccoli alpha-glucosidase indicated that it corresponds to the Arabidopsis Aglu-2 gene and that approximately 15 kD of the predicted N terminus was cleaved. The 81-kD protein was more abundant than the 96-kD protein, but it was not active with 4-methylumbelliferyl-alpha-D-glucopyranoside as the substrate and it did not bind to concanavalin A. In situ activity staining using 5-bromo-4-chloro-3-indolyl-alpha-D-glucopyranoside revealed that the acidic alpha-glucosidase activity is predominantly located in the outer cortex of broccoli stems and in vascular tissue, especially in leaf traces.
Figures



Similar articles
-
The gene glvA of Bacillus subtilis 168 encodes a metal-requiring, NAD(H)-dependent 6-phospho-alpha-glucosidase. Assignment to family 4 of the glycosylhydrolase superfamily.J Biol Chem. 1998 Oct 16;273(42):27347-56. doi: 10.1074/jbc.273.42.27347. J Biol Chem. 1998. PMID: 9765262
-
6-phospho-alpha-D-glucosidase from Fusobacterium mortiferum: cloning, expression, and assignment to family 4 of the glycosylhydrolases.J Bacteriol. 1997 Jul;179(13):4129-37. doi: 10.1128/jb.179.13.4129-4137.1997. J Bacteriol. 1997. PMID: 9209025 Free PMC article.
-
Aspartic proteinase genes in the Brassicaceae Arabidopsis thaliana and Brassica napus.Plant Mol Biol. 1997 Jan;33(1):187-92. doi: 10.1023/a:1005794917200. Plant Mol Biol. 1997. PMID: 9037171
-
The glucosinolate-degrading enzyme myrosinase in Brassicaceae is encoded by a gene family.Plant Mol Biol. 1992 Jan;18(2):387-98. doi: 10.1007/BF00034965. Plant Mol Biol. 1992. PMID: 1731996
-
Plant alpha-glucosidases of the glycoside hydrolase family 31. Molecular properties, substrate specificity, reaction mechanism, and comparison with family members of different origin.Plant Mol Biol. 1998 May;37(1):1-13. doi: 10.1023/a:1005925819741. Plant Mol Biol. 1998. PMID: 9620260 Review. No abstract available.
Cited by
-
Multiplex Fluorescent, Activity-Based Protein Profiling Identifies Active α-Glycosidases and Other Hydrolases in Plants.Plant Physiol. 2018 May;177(1):24-37. doi: 10.1104/pp.18.00250. Epub 2018 Mar 19. Plant Physiol. 2018. PMID: 29555787 Free PMC article.
-
A real-time fluorogenic assay for the visualization of glycoside hydrolase activity in planta.Plant Physiol. 2009 Dec;151(4):1741-50. doi: 10.1104/pp.109.147439. Epub 2009 Sep 25. Plant Physiol. 2009. PMID: 19783642 Free PMC article.
-
AXY3 encodes a α-xylosidase that impacts the structure and accessibility of the hemicellulose xyloglucan in Arabidopsis plant cell walls.Planta. 2011 Apr;233(4):707-19. doi: 10.1007/s00425-010-1330-7. Epub 2010 Dec 18. Planta. 2011. PMID: 21170548 Free PMC article.
-
MeGATAs, functional generalists in interactions between cassava growth and development, and abiotic stresses.AoB Plants. 2022 Nov 25;15(1):plac057. doi: 10.1093/aobpla/plac057. eCollection 2023 Jan. AoB Plants. 2022. PMID: 36654987 Free PMC article.
-
A sub-proteome of Arabidopsis thaliana mature stems trapped on Concanavalin A is enriched in cell wall glycoside hydrolases.J Exp Bot. 2007;58(10):2503-12. doi: 10.1093/jxb/erm082. Epub 2007 May 26. J Exp Bot. 2007. PMID: 17526915 Free PMC article.
References
-
- Arendt CW, Ostergaard HL. Identification of the CD45-associated 116-kDa and 80-kDa proteins as the alpha- and beta-subunits of alpha-glucosidase II. J Biol Chem. 1997;272:13117–13125. - PubMed
-
- Beckerman JL, Ebbole DJ. MPG1, a gene encoding a fungal hydrophobin of Magnaporthe grisea, is involved in surface recognition. Mol Plant Microbe Interact. 1996;9:450–456. - PubMed
-
- Chiba S, Inomata S, Matsui H, Shimomura T. Purification and properties of an α-glucosidase (glucoamylase) in sugar beet seed. Agric Biol Chem. 1978;42:241–245.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

