Molecular cloning and expression analysis of the mitochondrial pyruvate dehydrogenase from maize
- PMID: 9952460
- PMCID: PMC32141
- DOI: 10.1104/pp.119.2.635
Molecular cloning and expression analysis of the mitochondrial pyruvate dehydrogenase from maize
Abstract
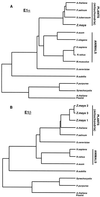
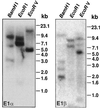
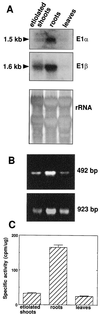
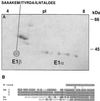
Four cDNAs, one encoding an alpha-subunit and three encoding beta-subunits of the mitochondrial pyruvate dehydrogenase, were isolated from maize (Zea mays L.) libraries. The deduced amino acid sequences of both alpha- and beta-subunits are approximately 80% identical with Arabidopsis and pea (Pisum sativum L.) homologs. The mature N terminus was determined for the beta-subunit by microsequencing the protein purified from etiolated maize shoot mitochondria and was resolved by two-dimensional gel electrophoresis. This single isoelectric species comprised multiple isoforms. Both alpha- and beta-subunits are encoded by multigene families in maize, as determined by Southern-blot analyses. RNA transcripts for both alpha- and beta-subunits were more abundant in roots than in young leaves or etiolated shoots. Pyruvate dehydrogenase activity was also higher in roots (5-fold) compared with etiolated shoots and leaves. Both subunits were present at similar levels in all tissues examined, indicating coordinated gene regulation. The protein levels were highest in heterotrophic organs and in pollen, which contained about 2-fold more protein than any other organ examined. The relative abundance of these proteins in nonphotosynthetic tissues may reflect a high cellular content of mitochondria, a high level of respiratory activity, or an extra plastidial requirement for acetate.
Figures




References
-
- Ali MS, Shenoy BC, Eswaran D, Andersson LA, Roche TE, Patel MS. Identification of the tryptophan residue in the thiamin pyrophosphate binding site of mammalian pyruvate dehydrogenase. J Biol Chem. 1995;270:4570–4574. - PubMed
-
- Eswaran D, Ali MS, Shenoy BC, Korotchkina LG, Roche TE, Patel MS. Arginine-239 in the beta subunit is at or near the active site of bovine pyruvate dehydrogenase. Biochim Biophys Acta. 1995;1252:203–208. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

