Functional interactions of the HHCC domain of moloney murine leukemia virus integrase revealed by nonoverlapping complementation and zinc-dependent dimerization
- PMID: 9971758
- PMCID: PMC104420
- DOI: 10.1128/JVI.73.3.1809-1817.1999
Functional interactions of the HHCC domain of moloney murine leukemia virus integrase revealed by nonoverlapping complementation and zinc-dependent dimerization
Abstract
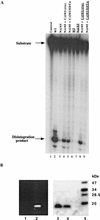
The retroviral integrase (IN) is required for the integration of viral DNA into the host genome. The N terminus of IN contains an HHCC zinc finger-like motif, which is conserved among all retroviruses. To study the function of the HHCC domain of Moloney murine leukemia virus IN, the first N-terminal 105 residues were expressed independently. This HHCC domain protein is found to complement a completely nonoverlapping construct lacking the HHCC domain for strand transfer, 3' processing and coordinated disintegration reactions, revealing trans interactions among IN domains. The HHCC domain protein binds zinc at a 1:1 ratio and changes its conformation upon binding to zinc. The presence of zinc within the HHCC domain stimulates selective integration processes. Zinc promotes the dimerization of the HHCC domain and protects it from N-ethylmaleimide modification. These studies dissect and define the requirement for the HHCC domain, the exact function of which remains unknown.
Figures

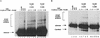



References
-
- Berg J M, Shi Y. The galvanization of biology: a growing appreciation for the roles of zinc. Science. 1996;271:1081–1085. - PubMed
-
- Berg J M. Zinc finger domains: hypotheses and current knowledge. Annu Rev Biophys Biophys Chem. 1990;19:405–421. - PubMed
-
- Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
-
- Brahms S, Brahms J. Determination of protein secondary structure in solution by vacuum ultraviolet circular dichroism. J Mol Biol. 1980;138:149–178. - PubMed
-
- Brown P O. Integration. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 161–203. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

