The chromatin structure of the long control region of human papillomavirus type 16 represses viral oncoprotein expression
- PMID: 9971771
- PMCID: PMC104433
- DOI: 10.1128/JVI.73.3.1918-1930.1999
The chromatin structure of the long control region of human papillomavirus type 16 represses viral oncoprotein expression
Abstract
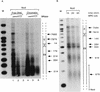
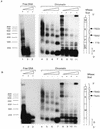
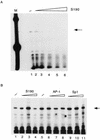
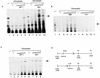
The long control region (LCR) of human papillomavirus type 16 (HPV-16) has a size of 850 bp (about 12% of the viral genome) and regulates transcription and replication of the viral DNA. The 5' segment of the LCR contains transcription termination signals and a nuclear matrix attachment region, the central segment contains an epithelial cell-specific enhancer, and the 3' segment contains the replication origin and the E6 promoter. Here we report observations on the chromatin organization of this part of the HPV-16 genome. Treatment of the nuclei of CaSki cells, a cell line with 500 intrachromosomal copies of HPV-16, with methidiumpropyl-EDTA-Fe(II) reveals nucleosomes in specific positions on the LCR and the E6 and E7 genes. One of these nucleosomes, which we termed Ne, overlaps with the center of the viral enhancer, while a second nucleosome, Np16, overlaps with the replication origin and the E6 promoter. The two nucleosomes become positioned on exactly the same segments after in vitro assembly of chromatin on the cloned HPV-16 LCR. Primer extension mapping of DNase I-cleaved chromatin revealed Np16 to be positioned centrally over E6 promoter elements, extending into the replication origin. Ne covers the center of the enhancer but leaves an AP-1 site, one of the strongest cis-responsive elements of the enhancer, unprotected. Np16, or a combination of Np16 and Ne, represses the activity of the E6 promoter during in vitro transcription of HPV-16 chromatin. Repression is relieved by addition of Sp1 and AP-1 transcription factors. Sp1 alters the structure of Np16 in vitro, while no changes can be observed during the binding of AP-1. HPV-18, which has a similar arrangement of cis-responsive elements despite its evolutionary divergence from HPV-16, shows specific assembly in vitro of a nucleosome, Np18, over the E1 binding site and E6 promoter elements but positioned about 90 bp 5' of the position of Np16 on the homologous HPV-16 sequences. The chromatin organization of the HPV-16 and HPV-18 genomes suggests important regulatory roles of nucleosomes during the viral life cycle.
Figures



References
-
- Bauer W R, Hayes J J, White J H, Wolffe A P. Nucleosome structural changes due to acetylation. J Mol Biol. 1994;236:685–690. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

