Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification
- PMID: 9971773
- PMCID: PMC104435
- DOI: 10.1128/JVI.73.3.1941-1948.1999
Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification
Abstract
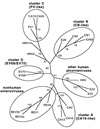
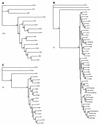
Sixty-six human enterovirus serotypes have been identified by serum neutralization, but the molecular determinants of the serotypes are unknown. Since the picornavirus VP1 protein contains a number of neutralization domains, we hypothesized that the VP1 sequence should correspond with neutralization (serotype) and, hence, with phylogenetic lineage. To test this hypothesis and to analyze the phylogenetic relationships among the human enteroviruses, we determined the complete VP1 sequences of the prototype strains of 47 human enterovirus serotypes and 10 antigenic variants. Our sequences, together with those available from GenBank, comprise a database of complete VP1 sequences for all 66 human enterovirus serotypes plus additional strains of seven serotypes. Phylogenetic trees constructed from complete VP1 sequences produced the same four major clusters as published trees based on partial VP2 sequences; in contrast to the VP2 trees, however, in the VP1 trees strains of the same serotype were always monophyletic. In pairwise comparisons of complete VP1 sequences, enteroviruses of the same serotype were clearly distinguished from those of heterologous serotypes, and the limits of intraserotypic divergence appeared to be about 25% nucleotide sequence difference or 12% amino acid sequence difference. Pairwise comparisons suggested that coxsackie A11 and A15 viruses should be classified as strains of the same serotype, as should coxsackie A13 and A18 viruses. Pairwise identity scores also distinguished between enteroviruses of different clusters and enteroviruses from picornaviruses of different genera. The data suggest that VP1 sequence comparisons may be valuable in enterovirus typing and in picornavirus taxonomy by assisting in the genus assignment of unclassified picornaviruses.
Figures

References
-
- Archarya R, Fry E, Stuart D, Fox G, Rowlands D J, Brown F. The three dimensional structure of foot and mouth disease virus at 2.9Å resolution. Nature. 1989;338:709–716. - PubMed
-
- Brown B A, Pallansch M A. Complete nucleotide sequence of enterovirus 71 is distinct from poliovirus. Virus Res. 1995;39:195–205. - PubMed
-
- Chang K, Auvinen P, Hyypiä T, Stanway G. The nucleotide sequence of coxsackievirus A9: implications for receptor binding and enterovirus classification. J Gen Virol. 1989;70:3269–3280. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

