Conditions for copackaging rous sarcoma virus and murine leukemia virus Gag proteins during retroviral budding
- PMID: 9971785
- PMCID: PMC104447
- DOI: 10.1128/JVI.73.3.2045-2051.1999
Conditions for copackaging rous sarcoma virus and murine leukemia virus Gag proteins during retroviral budding
Abstract
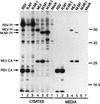
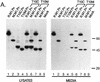
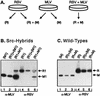
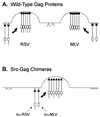
Rous sarcoma virus (RSV) and murine leukemia virus (MLV) are examples of distantly related retroviruses that normally do not encounter one another in nature. Their Gag proteins direct particle assembly at the plasma membrane but possess very little sequence similarity. As expected, coexpression of these two Gag proteins did not result in particles that contain both. However, when the N-terminal membrane-binding domain of each molecule was replaced with that of the Src oncoprotein, which is also targeted to the cytoplasmic face of the plasma membrane, efficient copackaging was observed in genetic complementation and coimmunoprecipitation assays. We hypothesize that the RSV and MLV Gag proteins normally use distinct locations on the plasma membrane for particle assembly but otherwise have assembly domains that are sufficiently similar in function (but not sequence) to allow heterologous interactions when these proteins are redirected to a common membrane location.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

