CD21-Dependent infection of an epithelial cell line, 293, by Epstein-Barr virus
- PMID: 9971794
- PMCID: PMC104456
- DOI: 10.1128/JVI.73.3.2115-2125.1999
CD21-Dependent infection of an epithelial cell line, 293, by Epstein-Barr virus
Abstract
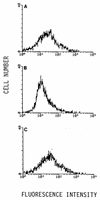
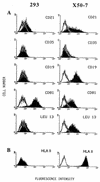
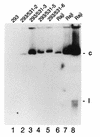
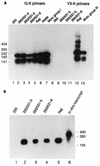
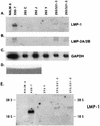
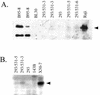
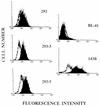
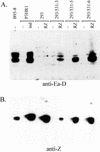
Epstein-Barr virus (EBV) is invariably present in undifferentiated nasopharyngeal carcinomas, is found sporadically in other carcinomas, and replicates in the differentiated layer of the tongue epithelium in lesions of oral hairy leukoplakia. However, it is not clear how frequently or by what mechanism EBV infects epithelial cells normally. Here, we report that a human epithelial cell line, 293, can be stably infected by EBV that has been genetically marked with a selectable gene. We show that 293 cells express a relatively low level of CD21, that binding of fluorescein-labeled EBV to 293 cells can be detected, and that both the binding of virus to cells and infection can be blocked with antibodies specific for CD21. Two proteins known to form complexes with CD21 on the surface of lymphoid cells, CD35 and CD19, could not be detected at the surface of 293 cells. All infected clones of 293 cells exhibited tight latency with a pattern of gene expression similar to that of type II latency, but productive EBV replication and release of infectious virus could be induced inefficiently by forced expression of the lytic transactivators, R and Z. Low levels of mRNA specific for the transforming membrane protein of EBV, LMP-1, as well as for LMP-2, were detected; however, LMP-1 protein was either undetectable or near the limit of detection at less than 5% of the level typical of EBV-transformed B cells. A slight increase in expression of the receptor for epidermal growth factor, which can be induced in epithelial cells by LMP-1, was detected at the cell surface with two EBV-infected 293 cell clones. These results show that low levels of surface CD21 can support infection of an epithelial cell line by EBV. The results also raise the possibility that in a normal infection of epithelial cells by EBV, the LMP-1 protein is not expressed at levels that are high enough to be oncogenic and that there might be differences in the cells of EBV-associated epithelial cancers that have arisen to allow for elevated expression of LMP-1.
Figures

References
-
- Anagnostopoulos I, Hummel M, Kreschel C, Stein H. Morphology, immunophenotype, and distribution of latently and/or productively Epstein-Barr virus-infected cells in acute infectious mononucleosis: implications for the interindividual infection route of Epstein-Barr virus. Blood. 1995;85:744–750. - PubMed
-
- Dawson C W, Rickinson A B, Young L S. Epstein-Barr virus latent membrane protein inhibits human epithelial cell differentiation. Nature. 1990;344:777–780. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

