Interclass transmission and phyletic host tracking in murine leukemia virus-related retroviruses
- PMID: 9971829
- PMCID: PMC104491
- DOI: 10.1128/JVI.73.3.2442-2449.1999
Interclass transmission and phyletic host tracking in murine leukemia virus-related retroviruses
Abstract
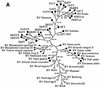
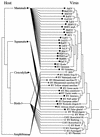
Retroviruses are capable of infectious horizontal transmission between hosts, usually between individuals within a single species, although a number of probable zoonotic infections resulting from transmission between different species of placental mammals have also been reported. Despite these data, it remains unclear how often interspecies transmission events occur or whether their frequency is influenced by the evolutionary distance between host taxa. To address this problem we used PCR to amplify and characterize endogenous retroviruses related to the murine leukemia viruses. We show that members of this retroviral genus are harbored by considerably more organisms than previously thought and that phylogenetic analysis demonstrates that viruses isolated from a particular host class generally cluster together, suggesting that infectious virus horizontal transfer between vertebrate classes occurs only rarely. However, two recent instances of transmission of zoonotic infections between distantly related host organisms were identified. One, from mammals to birds, has led to a rapid adaptive radiation into other avian hosts. The other, between placental and marsupial mammals, involves viruses clustering with recently described porcine retroviruses, adding to concerns regarding the xenotransplantation of pig organs to humans.
Figures


References
-
- Bach F H, Fishman J A, Daniels N, Proimos J, Anderson B, Carpenter C B, Forrow L, Robson S C, Fineberg H V. Uncertainty in xenotransplantation: individual benefit versus collective risk. Nat Med. 1998;4:141–142. - PubMed
-
- Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumor viruses. Nature. 1970;226:1209–1211. - PubMed
-
- Benveniste R E, Todaro G J. Evolution of C-type viral genes: inheritance of exogenously acquired viral genes. Nature. 1974;252:456–459. - PubMed
-
- Canfield P J, Brown A S, Kelly W R, Sutton R H. Spontaneous lymphoid neoplasia in the koala (Phascolarctos cinereus) J Comp Pathol. 1987;97:171–178. - PubMed
-
- Canfield P J, Sabine J M, Love D N. Virus particles associated with leukaemia in a koala. Aust Vet J. 1988;65:327–328. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

