Development of animal models for adeno-associated virus site-specific integration
- PMID: 9971837
- PMCID: PMC104499
- DOI: 10.1128/JVI.73.3.2517-2526.1999
Development of animal models for adeno-associated virus site-specific integration
Abstract
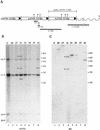
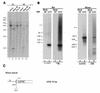
The adeno-associated virus (AAV) is unique in its ability to target viral DNA integration to a defined region of human chromosome 19 (AAVS1). Since AAVS1 sequences are not conserved in a rodent's genome, no animal model is currently available to study AAV-mediated site-specific integration. We describe here the generation of transgenic rats and mice that carry the AAVS1 3.5-kb DNA fragment. To test the response of the transgenic animals to Rep-mediated targeting, primary cultures of mouse fibroblasts, rat hepatocytes, and fibroblasts were infected with wild-type wt AAV. PCR amplification of the inverted terminal repeat (ITR)-AAVS1 junction revealed that the AAV genome integrated into the AAVS1 site in fibroblasts and hepatocytes. Integration in rat fibroblasts was also observed upon transfection of a plasmid containing the rep gene under the control of the p5 and p19 promoters and a dicistronic cassette carrying the green fluorescent protein (GFP) and neomycin (neo) resistance gene between the ITRs of AAV. The localization of the GFP-Neo sequence in the AAVS1 region was determined by Southern blot and FISH analysis. Lastly, AAV genomic DNA integration into the AAVS1 site in vivo was assessed by virus injection into the quadriceps muscle of transgenic rats and mice. Rep-mediated targeting to the AAVS1 site was detected in several injected animals. These results indicate that the transgenic lines are proficient for Rep-mediated targeting. These animals should allow further characterization of the molecular aspects of site-specific integration and testing of the efficacy of targeted integration of AAV recombinant vectors designed for human gene therapy.
Figures




Similar articles
-
Herpes simplex virus type 1/adeno-associated virus hybrid vectors mediate site-specific integration at the adeno-associated virus preintegration site, AAVS1, on human chromosome 19.J Virol. 2002 Jul;76(14):7163-73. doi: 10.1128/jvi.76.14.7163-7173.2002. J Virol. 2002. PMID: 12072516 Free PMC article.
-
Targeted transgene integration into transgenic mouse fibroblasts carrying the full-length human AAVS1 locus mediated by HSV/AAV rep(+) hybrid amplicon vector.Gene Ther. 2003 Sep;10(19):1691-702. doi: 10.1038/sj.gt.3302061. Gene Ther. 2003. PMID: 12923568
-
Site-specific integration of functional transgenes into the human genome by adeno/AAV hybrid vectors.Mol Ther. 2004 Oct;10(4):660-70. doi: 10.1016/j.ymthe.2004.07.003. Mol Ther. 2004. PMID: 15451450
-
Second generation adeno-associated virus type 2-based gene therapy systems with the potential for preferential integration into AAVS1.Curr Gene Ther. 2002 May;2(2):145-59. doi: 10.2174/1566523024605627. Curr Gene Ther. 2002. PMID: 12109212 Review.
-
Chimeric herpes simplex virus/adeno-associated virus amplicon vectors.Curr Gene Ther. 2006 Jun;6(3):315-24. doi: 10.2174/156652306777592090. Curr Gene Ther. 2006. PMID: 16787183 Review.
Cited by
-
Targeted Integration and High-Level Transgene Expression in AAVS1 Transgenic Mice after In Vivo HSC Transduction with HDAd5/35++ Vectors.Mol Ther. 2019 Dec 4;27(12):2195-2212. doi: 10.1016/j.ymthe.2019.08.006. Epub 2019 Aug 19. Mol Ther. 2019. PMID: 31494053 Free PMC article.
-
Characteristics of the adeno-associated virus preintegration site in human chromosome 19: open chromatin conformation and transcription-competent environment.J Virol. 2000 Aug;74(16):7671-7. doi: 10.1128/jvi.74.16.7671-7677.2000. J Virol. 2000. PMID: 10906224 Free PMC article.
-
Ribosomal DNA integrating rAAV-rDNA vectors allow for stable transgene expression.Mol Ther. 2012 Oct;20(10):1912-23. doi: 10.1038/mt.2012.164. Epub 2012 Sep 18. Mol Ther. 2012. PMID: 22990671 Free PMC article.
-
Adipose Tissue: An Emerging Target for Adeno-associated Viral Vectors.Mol Ther Methods Clin Dev. 2020 Sep 20;19:236-249. doi: 10.1016/j.omtm.2020.09.009. eCollection 2020 Dec 11. Mol Ther Methods Clin Dev. 2020. PMID: 33102616 Free PMC article. Review.
-
Identification and characterization of an adeno-associated virus integration site in CV-1 cells from the African green monkey.J Virol. 2003 Feb;77(3):1904-15. doi: 10.1128/jvi.77.3.1904-1915.2003. J Virol. 2003. PMID: 12525625 Free PMC article.
References
-
- Bartlett J S, Samulski J R. Methods in molecular medicine. In: Robbins P D, editor. Gene therapy protocols. Totowa, N.J: Humana Press, Inc.; 1997. pp. 25–40.
-
- Berns K I. Parvoviridiae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2173–2197.
-
- Boussif O, Zanta M A, Behr J-P. Optimized galenics improve in vitro gene transfer with cationic molecules up to 1000-fold. Gene Ther. 1996;3:1074–1080. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

