Purification, characterization, and cloning of a eubacterial 3-hydroxy-3-methylglutaryl coenzyme A reductase, a key enzyme involved in biosynthesis of terpenoids
- PMID: 9973353
- PMCID: PMC93504
- DOI: 10.1128/JB.181.4.1256-1263.1999
Purification, characterization, and cloning of a eubacterial 3-hydroxy-3-methylglutaryl coenzyme A reductase, a key enzyme involved in biosynthesis of terpenoids
Abstract
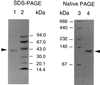
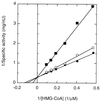
The eubacterial 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase (EC 1.1.1.34) was purified 3,000-fold from Streptomyces sp. strain CL190 to apparent homogeneity with an overall yield of 2.1%. The purification procedure consisted of (NH4)2SO4 precipitation, heat treatment and anion exchange, hydrophobic interaction, and affinity chromatographies. The molecular mass of the enzyme was estimated to be 41 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and 100 to 105 kDa by gel filtration chromatography, suggesting that the enzyme is most likely to be a dimer. The enzyme showed a pH optimum of around 7.2, with apparent Km values of 62 microM for NADPH and 7.7 microM for HMG-CoA. A gene from CL190 responsible for HMG-CoA reductase was cloned by the colony hybridization method with an oligonucleotide probe synthesized on the basis of the N-terminal sequence of the purified enzyme. The amino acid sequence of the CL190 HMG-CoA reductase revealed several limited motifs which were highly conserved and common to the eucaryotic and archaebacterial enzymes. These sequence conservations suggest a strong evolutionary pressure to maintain amino acid residues at specific positions, indicating that the conserved motifs might play important roles in the structural conformation and/or catalytic properties of the enzyme.
Figures

References
-
- Alberts A W, Chen J, Kuron G, Hunt V, Huff J, Hoffman C, Rothrock J, Lopez M, Joshua H, Harris E, Patchett A, Monaghan R, Currie S, Stapley E, Albers-Schonberg G, Hensens O, Hirshfield J, Hoogsteen K, Liesch J, Springer J. Mevinolin: a highly potent competitive inhibitor of hydroxymethylglutaryl-coenzyme A reductase and a cholesterol-lowering agent. Proc Natl Acad Sci USA. 1980;77:3957–3961. - PMC - PubMed
-
- Amemiya Y, Miyahara J. Imaging plate illuminates many fields. Nature. 1988;336:89–90. - PubMed
-
- Bach T J, Rogers D H, Rudney H. Detergent-solubilization, purification, and characterization of membrane-bound 3-hydroxy-3-methylglutaryl-coenzyme A reductase from radish seedlings. Eur J Biochem. 1986;154:103–111. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

