Auxins upregulate expression of the indole-3-pyruvate decarboxylase gene in Azospirillum brasilense
- PMID: 9973364
- PMCID: PMC93515
- DOI: 10.1128/JB.181.4.1338-1342.1999
Auxins upregulate expression of the indole-3-pyruvate decarboxylase gene in Azospirillum brasilense
Abstract
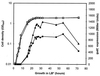
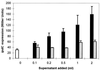
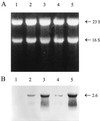
Transcription of the Azospirillum brasilense ipdC gene, encoding an indole-3-pyruvate decarboxylase involved in the biosynthesis of indole-3-acetic acid (IAA), is induced by IAA as determined by ipdC-gusA expression studies and Northern analysis. Besides IAA, exogenously added synthetic auxins such as 1-naphthaleneacetic acid, 2,4-dichlorophenoxypropionic acid, and p-chlorophenoxyacetic acid were also found to upregulate ipdC expression. No upregulation was observed with tryptophan, acetic acid, or propionic acid or with the IAA conjugates IAA ethyl ester and IAA-L-phenylalanine, indicating structural specificity is required for ipdC induction. This is the first report describing the induction of a bacterial gene by auxin.
Figures
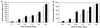
References
-
- Bainton N J, Bycroft B W, Chhabra S R, Stead P, Gledhill L, Hill P J, Rees C E D, Winson M K, Salmond G P C, Stewart G S A B, Williams P A. A general role for the lux autoinducer in bacterial cell signalling: control of antibiotic biosynthesis in Erwinia. Gene. 1992;116:87–91. - PubMed
-
- Bartel B. Auxin biosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:51–66. - PubMed
-
- Costacurta A, Keijers V, Vanderleyden J. Molecular cloning and sequence analysis of an Azospirillum brasilense indole-3-pyruvate decarboxylase gene. Mol Gen Genet. 1994;243:463–472. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

