Comparative analysis and serovar-specific identification of multiple-banded antigen genes of Ureaplasma urealyticum biovar 1
- PMID: 9986808
- PMCID: PMC84456
- DOI: 10.1128/JCM.37.3.538-543.1999
Comparative analysis and serovar-specific identification of multiple-banded antigen genes of Ureaplasma urealyticum biovar 1
Abstract
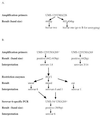
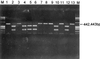
Ureaplasma urealyticum is a causative agent of nongonococcal urethritis and is implicated in the pathogenesis of several other diseases. The species is divided into 14 serovars and two biovars, of which biovar 1 is most commonly isolated from clinical specimens. Reported associations between individual serovars and diseases have been difficult to confirm because of practical difficulties with serotyping. The multiple-banded antigen (MBA) is the predominant U. urealyticum antigen recognized during infections in humans and probably has a significant role in virulence. The 5' end of the MBA gene is relatively conserved but contains biovar, and possibly serovar, specificity. The 5' ends of the MBA genes of standard strains of U. urealyticum biovar 1, consisting of serovars 1, 3, 6, and 14, were amplified, cloned into pUC19, and sequenced to identify serovar-specific differences. The 5' end of the MBA gene sequence of serovar 3 was identical with the previously published sequence and differed by only three bases from that of serovar 14. Significant differences between the MBA gene sequences allowed biovar 1 to be divided into two subgroups, containing serovars 3/14 and serovars 1 and 6, respectively, using primers UMS-125-UMA269 and UMS-125-UMA269'. Serovars 1 and 6 were distinguished by restriction enzyme analysis of the amplicon and/or by PCR specific for serovar 6. These methods were used to identify and type U. urealyticum in 185 (46.3%) of 400 genital specimens from women. Biovar 1 was detected in 89.2% and biovar 2 in 18.3% of positive specimens. Of 165 specimens containing U. urealyticum biovar 1, 22.2% contained more than one serovar and 46.7, 46.1, and 25.5% contained serovars 1, 3/14, and 6, respectively. U. urealyticum was found in a significantly higher proportion of pregnant women than in sex workers and other women attending a sexually transmissible diseases clinic (P < 0.01). The methods described are relatively rapid, practicable, and specific for serotyping isolates and for direct detection and identification of individual serovars in clinical specimens containing U. urealyticum biovar 1.
Figures




References
-
- Abele-Horn M, Peters J, Genzel-Boroviczeny O, Wolff C, Zimmermann A, Gottschling W. Vaginal Ureaplasma urealyticum colonization: influence on pregnancy outcome and neonatal morbidity. Infection. 1997;25:286–291. - PubMed
-
- Cassell G H, Crouse D T, Waites K B, Rudd P T, Davis J K. Does Ureaplasma urealyticum cause respiratory disease in newborns? Pediatr Infect Dis J. 1988;7:535–541. - PubMed
-
- Grattard F, Pozzetto B, de Barbeyrac B, Renaudin H, Clerc M, Gaudin O G, Bebear C. Arbitrarily-primed PCR confirms the differentiation of strains of Ureaplasma urealyticum to two biovars. Mol Cell Probes. 1995;9:383–389. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

