Genetic diversity and population structure of Vibrio cholerae
- PMID: 9986816
- PMCID: PMC84478
- DOI: 10.1128/JCM.37.3.581-590.1999
Genetic diversity and population structure of Vibrio cholerae
Abstract
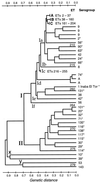
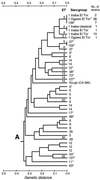
Multilocus enzyme electrophoresis (MLEE) of 397 Vibrio cholerae isolates, including 143 serogroup reference strains and 244 strains from Mexico and Guatemala, identified 279 electrophoretic types (ETs) distributed in two major divisions (I and II). Linkage disequilibrium was demonstrated in both divisions and in subdivision Ic of division I but not in subdivision Ia, which includes 76% of the ETs. Despite this evidence of relatively frequent recombination, clonal lineages may persist for periods of time measured in at least decades. In addition to the pandemic clones of serogroups O1 and O139, which form a tight cluster of four ETs in subdivision Ia, MLEE analysis identified numerous apparent clonal lineages of non-O1 strains with intercontinental distributions. A clone of serogroup O37 that demonstrated epidemic potential in the 1960s is closely related to the pandemic O1/O139 clones, but the nontoxigenic O1 Inaba El Tor reference strain is not. A strain of serogroup O22, which has been identified as the most likely donor of exogenous rfb region DNA to the O1 progenitor of the O139 clone, is distantly related to the O1/O139 clones. The close evolutionary relationships of the O1, O139, and O37 epidemic clones indicates that new cholera clones are likely to arise by the modification of a lineage that is already epidemic or is closely related to such a clone.
Figures
References
-
- Albert M J, Siddique A K, Islam M S, Faruque A S, Ansaruzzaman M, Faruque S M, Sack R B. Large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet. 1993;341:704. - PubMed
-
- Blake P A, Weaver R E, Hollis D G. Diseases of humans (other than cholera) caused by vibrios. Annu Rev Microbiol. 1980;34:341–367. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

