Improved silica-guanidiniumthiocyanate DNA isolation procedure based on selective binding of bovine alpha-casein to silica particles
- PMID: 9986822
- PMCID: PMC84491
- DOI: 10.1128/JCM.37.3.615-619.1999
Improved silica-guanidiniumthiocyanate DNA isolation procedure based on selective binding of bovine alpha-casein to silica particles
Abstract
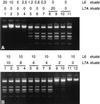
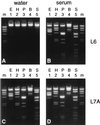
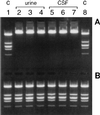
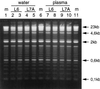
DNA purified from clinical cerebrospinal fluid and urine specimens by a silica-guanidiniumthiocyanate procedure frequently contained an inhibitor(s) of DNA-processing enzymes which may have been introduced by the purification procedure itself. Inhibition could be relieved by the use of a novel lysis buffer containing alpha-casein. When the novel lysis buffer was used, alpha-casein was bound by the silica particles in the first step of the procedure and eluted together with DNA in the last step, after which it exerted its beneficial effects for DNA-processing enzymes. In the present study we have compared the novel lysis buffer with the previously described lysis buffer with respect to double-stranded DNA yield (which was nearly 100%) and the performance of DNA-processing enzymes.
Figures
References
-
- Aaij C, Borst P. The gel electrophoresis of DNA. Biochim Biophys Acta. 1972;269:192–200. - PubMed
-
- Akrigg A, Wilkinson G W G, Oram J D. The structure of the major immediate early gene of human cytomegalovirus strain AD169. Virus Res. 1985;2:107–121. - PubMed
-
- Bernos E, Girardet J M, Humbert G, Linden G. Role of the O-phosphoserine clusters in the interaction of the bovine milk alpha s1-, beta-, kappa-caseins and the PP3 component with immobilized iron (III) ions. Biochim Biophys Acta. 1997;1337:149–159. - PubMed
-
- Blum H, Beier H, Gross H J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–98.
-
- Boom, R. Unpublished data.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

