Validation of binary typing for Staphylococcus aureus strains
- PMID: 9986829
- PMCID: PMC84511
- DOI: 10.1128/JCM.37.3.664-674.1999
Validation of binary typing for Staphylococcus aureus strains
Abstract
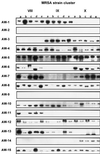
Most of the DNA-based methods for genetic typing of Staphylococcus aureus strains generate complex banding patterns. Therefore, we have developed a binary typing procedure involving strain-differentiating DNA probes which were generated on the basis of randomly amplified polymorphic DNA (RAPD) analysis. We present and validate the usefulness of 15 DNA probes, according to generally accepted performance criteria for molecular typing systems. RAPD analysis with multiple primers was performed on 376 S. aureus strains of which 97% were methicillin resistant (MRSA). Among the 1,128 RAPD patterns generated, 66 were selected which identified 124 unique DNA fragments. From these amplicons, only 12% turned out to be useful for isolate-specific binary typing. The nature of the RAPD-generated DNA fragments was investigated by partial DNA sequence analysis. Several homologies with known S. aureus sequences and with genes from other species were discovered; however, 87% of the probe sequences are of previously unknown origin. The locations of most of the DNA probes on the chromosome of S. aureus NCTC 8325 were determined by hybridization. Seven fragments were randomly dispersed along the genome, five were clustered within the 2500- to 2600-kb position of the genome, and the remaining four did not recognize complementary sequences in S. aureus NCTC 8325. A total of 103 S. aureus strains (69% MRSA) were used for the validation of the binary typing technique. The 15 DNA probes provided stable epidemiological markers, both in vitro (type consistency after serial passages on culture media) and in vivo (comparison of sequential isolates recovered from cases of persistent colonization). The discriminatory power of binary typing (D = 0.998) exceeded that of pulsed-field gel electrophoresis (D = 0.966) and RAPD analysis (D = 0.949). Reproducibility, measured by analyzing multiple strains belonging to a multitude of different epidemiological clusters, was comparable to that of other genotyping techniques used. Contribution of the DNA probes to the discriminatory power of the system was analyzed by comparison of dendrograms. This study demonstrates that binary typing is a robust tool for the genetic typing of S. aureus isolates.
Figures


References
-
- Aires de Sousa M, Sanches I S, van Belkum A, van Leeuwen W, Verbrugh H, de Lencastre H. Characterization of methicillin-resistant Staphylococcus aureus isolates from Portuguese hospitals by multiple genotyping systems. Microb Drug Res. 1996;2:331–341. - PubMed
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Arbeit R D. Laboratory procedures for the epidemiologic analysis of staphylococci. In: Archer G L, Crossley K B, editors. Staphylococci and staphylococcal diseases. New York, N.Y: Churchill Livingstone; 1998. pp. 203–286.
-
- Ayliffe G A J. The progressive intercontinental spread of methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 1997;24:S74–S79. - PubMed
-
- Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

