hsp65 sequencing for identification of rapidly growing mycobacteria
- PMID: 9986875
- PMCID: PMC84584
- DOI: 10.1128/JCM.37.3.852-857.1999
hsp65 sequencing for identification of rapidly growing mycobacteria
Abstract
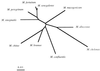
Partial sequencing of the hsp65 gene was used for the identification of rapidly growing mycobacteria (RGM). A 441-bp fragment (A. Telenti, F. Marchesi, M. Balz, F. Bally, E. Böttger, and T. Bodmer, J. Clin. Microbiol. 31:175-178, 1993) was amplified and sequenced by an automated fluorescence-based method involving capillary electrophoresis. Type strains of 10 RGM species were first studied. Each species had a unique nucleotide sequence, distinguishing it clearly from the other species. A panel of strains from the four main RGM species responsible for human infections, Mycobacterium abscessus, Mycobacterium chelonae, Mycobacterium fortuitum, and Mycobacterium peregrinum, was also studied. There were few sequence differences within each of these species (<2% of bases were different from the type strain sequence), and they had no effect on species assignment. hsp65 sequencing unambiguously differentiated M. chelonae and M. abscessus, two species difficult to identify by classical methods and 16S rRNA gene sequencing. The devised procedure is a rapid and reliable tool for the identification of RGM species.
Figures

References
-
- Hance A J, Grandchamp B, Lévy-Frébault V, Lecossier D, Rauzier J, Bocart D, Gicquel B. Detection and identification of mycobacteria by amplification of mycobacterial DNA. Mol Microbiol. 1989;3:843–849. - PubMed
-
- Ingram C W, Tanner D C, Durack D T, Kernodle G W, Jr, Corey G R. Disseminated infection with rapidly growing mycobacteria. Clin Infect Dis. 1993;16:463–471. - PubMed
-
- Kapur V, Li L L, Hamrick M R, Plikaytis B B, Shinnick T M, Telenti A, Jacobs W R, Banerjee A, Cole S, Yuen K Y, Clarridge J E, Kreiswirth B N, Musser J M. Rapid Mycobacterium species assignment and unambiguous identification of mutations associated with antimicrobial resistance in Mycobacterium tuberculosis by automated DNA sequencing. Arch Pathol Lab Med. 1995;119:131–138. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

