A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction
- PMID: 9990033
- PMCID: PMC15472
- DOI: 10.1073/pnas.96.4.1385
A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction
Abstract
Although present in many copies in the mouse genome, xenotropic murine leukemia viruses cannot infect cells from laboratory mice because of the lack of a functional cell surface receptor required for virus entry. In contrast, cells from many nonmurine species, including human cells, are fully permissive. Using an expression library approach, we isolated a cDNA from HeLa cell RNA that conferred susceptibility to xenotropic envelope protein binding and virus infection when expressed in nonpermissive cells. The deduced product is a 696-aa multiple-membrane spanning molecule, is widely expressed in human tissues, and shares homology with nematode, fly, and plant proteins of unknown function as well as with the yeast SYG1 protein, which has been shown to interact with a G protein. This molecule also acts as a receptor for polytropic murine leukemia viruses, consistent with observed interference between xenotropic and polytropic viruses in some cell types. This xenotropic and polytropic retrovirus receptor (XPR1) is the fourth identified molecule having multiple membrane spanning domains among mammalian type C oncoretrovirus receptors and may play a role in G protein-coupled signal transduction, as do the chemokine receptors required for HIV entry.
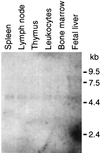
Figures



References
-
- Weiss R A. In: The Retroviridae. Levy J A, editor. Vol. 2. New York: Plenum; 1993. pp. 1–108.
-
- Hunter E. In: Retroviruses. Coffin J M, Hughes S H, Varmus H E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 71–119. - PubMed
-
- Sommerfelt M A, Weiss R A. Virology. 1990;176:58–69. - PubMed
-
- Patience C, Takeuchi Y, Weiss R A. Nat Med. 1997;3:282–286. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

