Binding of SARS-CoV-1/2 NSP1 to DNA Polymerase α-Primase Inhibits DNA Replication through Reduction of Interaction between DNA and DNA Polymerase α-Primase
- PMID: 40737240
- PMCID: PMC12344767
- DOI: 10.1021/acs.jcim.5c00999
Binding of SARS-CoV-1/2 NSP1 to DNA Polymerase α-Primase Inhibits DNA Replication through Reduction of Interaction between DNA and DNA Polymerase α-Primase
Abstract
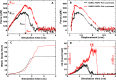
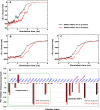
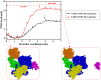
A comprehensive understanding of the atomic level mechanism governing the binding nonstructural protein 1 of SARS-CoV-1 (SARS-CoV-1 NSP1) and SARS-CoV-2 (SARS-CoV-2 NSP1) to Pol α-primase is important to advance the development of small molecule inhibitors for the treatment COVID-19. In this study, we use both all-atom steered molecular dynamics (all-atom SMD) and coarse-grained umbrella sampling (coarse-grained US) simulations to assess the binding affinity of SARS-CoV-1 NSP1 and SARS-CoV-2 NSP1 to Pol α-primase. Our all-atom SMD and coarse-grained US simulations consistently indicate that SARS-CoV-2 NSP1 exhibits stronger affinity for Pol α-primase compared to SARS-CoV-1 NSP1, implying that SARS-CoV-2 poses a greater risk than SARS-CoV-1 in impeding DNA replication for DNA synthesis. Through an energetic decomposition analysis of the interaction energies within these complexes, we identify electrostatic interactions as the primary contributors to the observed difference in binding affinity. We found that hydrogen bonds between Asp33 and Arg616 in SARS1 NSP1-Pol α-primase, and Asp33 with Arg616 and Lys655 in SARS2 NSP1-Pol α-primase, are critical for the interaction of both SARS-CoV-1 NSP1 and SARS-CoV-2 NSP1 with Pol α-primase. Asp33 in SARS-CoV-2 NSP1 shows increased solubility and stability compared to SARS-CoV-1 NSP1, enhancing its association with Pol α-primase. This finding lays the groundwork for innovative strategies aimed at inhibiting the interaction between these entities, offering promising avenues for therapeutic intervention against COVID-19. We also estimated the binding free energy of DNA to Pol α-primase, SARS1 NSP1-Pol α-primase, and SARS2 NSP1-Pol α-primase using the MM-PBSA method. The results show the order: Pol α-primase-DNA < SARS1 NSP1-Pol α-primase-DNA < SARS2 NSP1-Pol α-primase-DNA, indicating that both SARS-CoV-1 NSP1 and SARS-CoV-2 NSP1 reduce DNA binding to Pol α-primase, suggesting impaired DNA synthesis.
Figures

Similar articles
-
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Dec 16;12(12):CD014780. doi: 10.1002/14651858.CD014780. Cochrane Database Syst Rev. 2024. PMID: 39679851 Free PMC article.
-
Workplace interventions to reduce the risk of SARS-CoV-2 infection outside of healthcare settings.Cochrane Database Syst Rev. 2022 May 6;5(5):CD015112. doi: 10.1002/14651858.CD015112.pub2. Cochrane Database Syst Rev. 2022. Update in: Cochrane Database Syst Rev. 2024 Apr 10;4:CD015112. doi: 10.1002/14651858.CD015112.pub3. PMID: 35514111 Free PMC article. Updated.
-
The segmented flavivirus Alongshan virus reduces mitochondrial mass by degrading STAT2 to suppress the innate immune response.J Virol. 2025 Jan 31;99(1):e0130124. doi: 10.1128/jvi.01301-24. Epub 2024 Dec 10. J Virol. 2025. PMID: 39655955 Free PMC article.
-
Measures implemented in the school setting to contain the COVID-19 pandemic.Cochrane Database Syst Rev. 2022 Jan 17;1(1):CD015029. doi: 10.1002/14651858.CD015029. Cochrane Database Syst Rev. 2022. Update in: Cochrane Database Syst Rev. 2024 May 2;5:CD015029. doi: 10.1002/14651858.CD015029.pub2. PMID: 35037252 Free PMC article. Updated.
-
Structure-function mapping and mechanistic insights on the SARS CoV2 Nsp1.Protein Sci. 2024 Dec;33(12):e5228. doi: 10.1002/pro.5228. Protein Sci. 2024. PMID: 39584680
References
-
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H. L., Peiris M., Wu J.. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. - DOI - PMC - PubMed
-
- Wu F., Zhao S., Yu B., Chen Y. M., Wang W., Song Z. G., Hu Y., Tao Z. W., Tian J. H., Pei Y. Y., Yuan M. L., Zhang Y. L., Dai F. H., Liu Y., Wang Q. M., Zheng J. J., Xu L., Holmes E. C., Zhang Y. Z.. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. - DOI - PMC - PubMed
-
- Zhou P., Yang X. L., Wang X. G., Hu B., Zhang L., Zhang W., Si H. R., Zhu Y., Li B., Huang C. L., Chen H. D., Chen J., Luo Y., Guo H., Jiang R. D., Liu M. Q., Chen Y., Shen X. R., Wang X., Zheng X. S., Zhao K., Chen Q. J., Deng F., Liu L. L., Yan B., Zhan F. X., Wang Y. Y., Xiao G. F., Shi Z. L.. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

