Colorimetric RT-LAMP for SARS-CoV-2 detection from nasopharyngeal swabs or crude saliva: a multicountry diagnostic accuracy study in Africa
- PMID: 40580991
- PMCID: PMC12208784
- DOI: 10.1016/S2214-109X(25)00150-0
Colorimetric RT-LAMP for SARS-CoV-2 detection from nasopharyngeal swabs or crude saliva: a multicountry diagnostic accuracy study in Africa
Abstract
Background: Reverse-transcription loop-mediated isothermal amplification (RT-LAMP) with colorimetric readout is a rapid, robust, and cost-effective one-step amplification assay that we previously trialled for the identification of SARS-CoV-2 in nasopharyngeal swabs in four countries. Here, we expanded our assessment of RT-LAMP for SARS-CoV-2 detection to several other African countries and evaluated its operational performance with crude saliva as a pragmatic approach for outbreak surveillance and response in resource-limited settings.
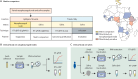
Methods: We conducted a multicountry diagnostic accuracy study of RT-LAMP for the detection of SARS-CoV-2 in different types of clinical samples. A preliminary study was conducted in Slovenia and Italy to establish the analytical performance (limit of detection) of RT-LAMP and optimise this assay before its deployment in Africa. Subsequently, we tested RT-LAMP with RNA extracted from nasopharyngeal swabs in seven countries in Africa (Angola, Burkina Faso, Côte d'Ivoire, Ethiopia, Senegal, Sudan, and Zimbabwe), and, in parallel, with crude saliva samples (ie, without RNA extraction) in an additional four countries (Cameroon, Ethiopia, Kenya, and Nigeria; paired nasopharyngeal swabs were collected at the same time). In both contexts, quantitative RT-PCR (RT-qPCR) with RNA extracted from nasopharyngeal swabs was used as the gold-standard benchmarking assay to evaluate performance. For RT-qPCR testing, each laboratory followed their own standard diagnostic procedure, whereas a standardised protocol was used for RT-LAMP. Saliva test standardisation was ensured through centralised reagent distribution. We calculated diagnostic parameters (sensitivity, specificity, and accuracy) using a 2 × 2 contingency table.
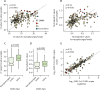
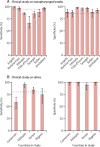
Findings: The preliminary study reported 87% sensitivity and 98% specificity for RT-LAMP. Between Sept 1, 2021, and June 30, 2022, we collected 2774 nasopharyngeal swabs and 577 crude saliva samples. For RNA extracted from nasopharyngeal swabs, the sensitivity and specificity of RT-LAMP for detection of SARS-CoV-2 (relative to the standard of diagnostics-ie, the RT-qPCR assay used in each participating laboratory) were 89% (95% CI 87-90) and 95% (93-96), respectively. Similarly, RT-LAMP tested on saliva without RNA extraction showed 80% (75-84) sensitivity and 99% (96-100) specificity (relative to the results obtained with the standard of diagnostics for RNA extracted from paired nasopharyngeal samples).
Interpretation: Colorimetric RT-LAMP is a reliable assay for SARS-CoV-2 detection in both extracted RNA and crude saliva samples. The demonstrably acceptable performance on crude saliva samples (without RNA extraction) underscores the scalability of this method for efficient outbreak surveillance in resource-limited settings.
Funding: Gates Foundation.
Copyright © 2025 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY 4.0 license. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of interests We declare no competing interests.
Figures

Similar articles
-
PD-LAMP smartphone detection of SARS-CoV-2 on chip.Anal Chim Acta. 2022 Apr 22;1203:339702. doi: 10.1016/j.aca.2022.339702. Epub 2022 Mar 9. Anal Chim Acta. 2022. PMID: 35361434 Free PMC article.
-
Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2022 Jul 22;7(7):CD013705. doi: 10.1002/14651858.CD013705.pub3. Cochrane Database Syst Rev. 2022. PMID: 35866452 Free PMC article.
-
An alternative real-time fluorescence reverse transcription loop-mediated isothermal amplification assay for the rapid detection of SARS-CoV-2.Rev Inst Med Trop Sao Paulo. 2025 Jun 27;67:e37. doi: 10.1590/S1678-9946202567037. eCollection 2025. Rev Inst Med Trop Sao Paulo. 2025. PMID: 40608622 Free PMC article.
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
References
-
- WHO Coronavirus disease (COVID-19 pandemic) https://www.who.int/emergencies/diseases/novel-coronavirus-2019
-
- Ranney ML, Griffeth V, Jha AK. Critical supply shortages—the need for ventilators and personal protective equipment during the Covid-19 pandemic. N Engl J Med. 2020;382:e41. - PubMed
-
- Sahoo A, Shulania A, Chhabra M, et al. Performance of chip based real time RT-PCR (TrueNat) and conventional real time RT-PCR for detection of SARS-CoV-2. J Clin of Diagn Res. 2021;15:DC25–DC28.
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

