High-Resolution Linear Epitope Mapping of the Receptor Binding Domain of SARS-CoV-2 Spike Protein in COVID-19 mRNA Vaccine Recipients
- PMID: 34756082
- PMCID: PMC8579840
- DOI: 10.1128/Spectrum.00965-21
High-Resolution Linear Epitope Mapping of the Receptor Binding Domain of SARS-CoV-2 Spike Protein in COVID-19 mRNA Vaccine Recipients
Abstract
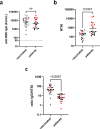
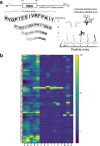
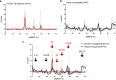
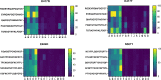
The prompt rollout of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) mRNA vaccine is facilitating population immunity, which is becoming more dominant than natural infection-mediated immunity. In the midst of coronavirus disease 2019 (COVID-19) vaccine deployment, understanding the epitope profiles of vaccine-elicited antibodies will be the first step in assessing the functionality of vaccine-induced immunity. In this study, the high-resolution linear epitope profiles of Pfizer-BioNTech COVID-19 mRNA vaccine recipients and COVID-19 patients were delineated by using microarrays mapped with overlapping peptides of the receptor binding domain (RBD) of the SARS-CoV-2 spike protein. The vaccine-induced antibodies targeting the RBD had a broader distribution across the RBD than that induced by the natural infection. Half-maximal neutralization titers were measured in vitro by live virus neutralization assays. As a result, relatively lower neutralizability was observed in vaccine recipient sera, when normalized to a total anti-RBD IgG titer. However, mutation panel assays targeting the SARS-CoV-2 variants of concern have shown that the vaccine-induced epitope variety, rich in breadth, may grant resistance against future viral evolutionary escapes, serving as an advantage of vaccine-induced immunity. IMPORTANCE Establishing vaccine-based population immunity has been the key factor in attaining herd protection. Thanks to expedited worldwide research efforts, the potency of mRNA vaccines against the coronavirus disease 2019 (COVID-19) is now incontestable. The next debate is regarding the coverage of SARS-CoV-2 variants. In the midst of vaccine deployment, it is of importance to describe the similarities and differences between the immune responses of COVID-19 vaccine recipients and naturally infected individuals. In this study, we demonstrated that the antibody profiles of vaccine recipients are richer in variety, targeting a key protein of the invading virus, than those of naturally infected individuals. Vaccine-elicited antibodies included more nonneutralizing antibodies than infection-elicited antibodies, and their breadth in antibody variations suggested possible resilience against future SARS-CoV-2 variants. The antibody profile achieved by vaccinations in naive individuals provides important insight into the first step toward vaccine-based population immunity.
Keywords: COVID-19; RBD; SARS-CoV-2; immunoserology; neutralizing antibodies; serology; spike; spike protein.
Conflict of interest statement
We declare no conflicts of interest.
Figures


Similar articles
-
Comprehensive characterization of the antibody responses to SARS-CoV-2 Spike protein finds additional vaccine-induced epitopes beyond those for mild infection.Elife. 2022 Jan 24;11:e73490. doi: 10.7554/eLife.73490. Elife. 2022. PMID: 35072628 Free PMC article.
-
Early 2022 breakthrough infection sera from India target the conserved cryptic class 5 epitope to counteract immune escape by SARS-CoV-2 variants.J Virol. 2025 Apr 15;99(4):e0005125. doi: 10.1128/jvi.00051-25. Epub 2025 Mar 26. J Virol. 2025. PMID: 40135898 Free PMC article.
-
SARS-CoV-2 mRNA vaccine induces robust specific and cross-reactive IgG and unequal neutralizing antibodies in naive and previously infected people.Cell Rep. 2022 Feb 1;38(5):110336. doi: 10.1016/j.celrep.2022.110336. Epub 2022 Jan 20. Cell Rep. 2022. PMID: 35090596 Free PMC article.
-
A Structural Landscape of Neutralizing Antibodies Against SARS-CoV-2 Receptor Binding Domain.Front Immunol. 2021 Apr 28;12:647934. doi: 10.3389/fimmu.2021.647934. eCollection 2021. Front Immunol. 2021. PMID: 33995366 Free PMC article. Review.
-
Severe acute respiratory syndrome-coronavirus-2 spike (S) protein based vaccine candidates: State of the art and future prospects.Rev Med Virol. 2021 May;31(3):e2183. doi: 10.1002/rmv.2183. Epub 2020 Oct 15. Rev Med Virol. 2021. PMID: 33594794 Free PMC article. Review.
Cited by
-
Omicron: a drug developer's perspective.Emerg Microbes Infect. 2022 Dec;11(1):208-211. doi: 10.1080/22221751.2021.2023330. Emerg Microbes Infect. 2022. PMID: 34951568 Free PMC article.
-
Heparan sulfate proteoglycans remodel SARS-CoV-2 spike conformation to allow integrin interaction and infection of endothelial cells.Front Cell Infect Microbiol. 2025 Apr 3;15:1552116. doi: 10.3389/fcimb.2025.1552116. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40248367 Free PMC article.
-
Immunodominant antibody responses directed to SARS-CoV-2 hotspot mutation sites and risk of immune escape.Front Immunol. 2023 Jan 5;13:1010105. doi: 10.3389/fimmu.2022.1010105. eCollection 2022. Front Immunol. 2023. PMID: 36685521 Free PMC article.
-
Antigen-adjuvant interactions, stability, and immunogenicity profiles of a SARS-CoV-2 receptor-binding domain (RBD) antigen formulated with aluminum salt and CpG adjuvants.Hum Vaccin Immunother. 2022 Nov 30;18(5):2079346. doi: 10.1080/21645515.2022.2079346. Epub 2022 Jun 6. Hum Vaccin Immunother. 2022. PMID: 35666264 Free PMC article.
-
Kinetics of anti-SARS-CoV-2 antibody titer in healthy adults up to 6 months after BNT162b2 vaccination measured by two immunoassays: A prospective cohort study in Japan.Vaccine. 2022 Sep 9;40(38):5631-5640. doi: 10.1016/j.vaccine.2022.08.018. Epub 2022 Aug 15. Vaccine. 2022. PMID: 36028457 Free PMC article.
References
-
- Ebinger JE, Fert-Bober J, Printsev I, Wu M, Sun N, Prostko JC, Frias EC, Stewart JL, Van Eyk JE, Braun JG, Cheng S, Sobhani K. 2021. Antibody responses to the BNT162b2 mRNA vaccine in individuals previously infected with SARS-CoV-2. Nat Med 27:981–984. doi:10.1038/s41591-021-01325-6. - DOI - PMC - PubMed
-
- Walsh EE, Frenck RW, Falsey AR, Kitchin N, Absalon J, Gurtman A, Lockhart S, Neuzil K, Mulligan MJ, Bailey R, Swanson KA, Li P, Koury K, Kalina W, Cooper D, Fontes-Garfias C, Shi P-Y, Türeci Ö, Tompkins KR, Lyke KE, Raabe V, Dormitzer PR, Jansen KU, Şahin U, Gruber WC. 2020. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N Engl J Med 383:2439–2450. doi:10.1056/NEJMoa2027906. - DOI - PMC - PubMed
-
- Mulligan MJ, Lyke KE, Kitchin N, Absalon J, Gurtman A, Lockhart S, Neuzil K, Raabe V, Bailey R, Swanson KA, Li P, Koury K, Kalina W, Cooper D, Fontes-Garfias C, Shi PY, Türeci Ö, Tompkins KR, Walsh EE, Frenck R, Falsey AR, Dormitzer PR, Gruber WC, Şahin U, Jansen KU. 2020. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 586:589–593. doi:10.1038/s41586-020-2639-4. - DOI - PubMed
-
- Müller L, Andrée M, Moskorz W, Drexler I, Walotka L, Grothmann R, Ptok J, Hillebrandt J, Ritchie A, Rabl D, Ostermann PN, Robitzsch R, Hauka S, Walker A, Menne C, Grutza R, Timm J, Adams O, Schaal H. 2021. Age-dependent immune response to the Biontech/Pfizer BNT162b2 COVID-19 vaccination. medRxiv 10.1101/2021.03.03.21251066. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- JP20wm0125003/Japan Agency for Medical Research and Development (AMED)
- JP20jk0110021/Japan Agency for Medical Research and Development (AMED)
- JP21441824/MEXT | Japan Society for the Promotion of Science (JSPS)
- Special Reserves fund for COVID-19/Osaka City University (OCU)
- COVID-19 Private Fund from the Shinya Yamanaka laboratory/Center for iPS Cell Research and Application, Kyoto University
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

