Comparison of Compression-Based Measures with Application to the Evolution of Primate Genomes
- PMID: 33265483
- PMCID: PMC7512912
- DOI: 10.3390/e20060393
Comparison of Compression-Based Measures with Application to the Evolution of Primate Genomes
Abstract
An efficient DNA compressor furnishes an approximation to measure and compare information quantities present in, between and across DNA sequences, regardless of the characteristics of the sources. In this paper, we compare directly two information measures, the Normalized Compression Distance (NCD) and the Normalized Relative Compression (NRC). These measures answer different questions; the NCD measures how similar both strings are (in terms of information content) and the NRC (which, in general, is nonsymmetric) indicates the fraction of one of them that cannot be constructed using information from the other one. This leads to the problem of finding out which measure (or question) is more suitable for the answer we need. For computing both, we use a state of the art DNA sequence compressor that we benchmark with some top compressors in different compression modes. Then, we apply the compressor on DNA sequences with different scales and natures, first using synthetic sequences and then on real DNA sequences. The last include mitochondrial DNA (mtDNA), messenger RNA (mRNA) and genomic DNA (gDNA) of seven primates. We provide several insights into evolutionary acceleration rates at different scales, namely, the observation and confirmation across the whole genomes of a higher variation rate of the mtDNA relative to the gDNA. We also show the importance of relative compression for localizing similar information regions using mtDNA.
Keywords: DNA sequences; NCD; NRC; data compression; primate evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
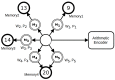
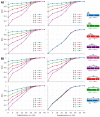


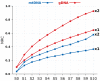
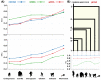

References
-
- Kolmogorov A.N. Three approaches to the quantitative definition of information. Probl. Inf. Transm. 1965;1:1–7. doi: 10.1080/00207166808803030. - DOI
-
- Niven R.K. Combinatorial entropies and statistics. Eur. Phys. J. B. 2009;70:49–63. doi: 10.1140/epjb/e2009-00168-5. - DOI
-
- Mantaci S., Restivo A., Rosone G., Sciortino M. A new combinatorial approach to sequence comparison. Theory Comput. Syst. 2008;42:411–429. doi: 10.1007/s00224-007-9078-6. - DOI
-
- Shannon C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948;27:379–423, 623–656. doi: 10.1002/j.1538-7305.1948.tb01338.x. - DOI
-
- Solomonoff R.J. A formal theory of inductive inference. Part I. Inf. Control. 1964;7:1–22. doi: 10.1016/S0019-9958(64)90223-2. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

