Analysis of xylem formation in pine by cDNA sequencing
- PMID: 9689143
- PMCID: PMC21401
- DOI: 10.1073/pnas.95.16.9693
Analysis of xylem formation in pine by cDNA sequencing
Abstract
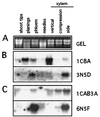
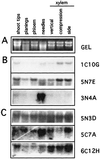
Secondary xylem (wood) formation is likely to involve some genes expressed rarely or not at all in herbaceous plants. Moreover, environmental and developmental stimuli influence secondary xylem differentiation, producing morphological and chemical changes in wood. To increase our understanding of xylem formation, and to provide material for comparative analysis of gymnosperm and angiosperm sequences, ESTs were obtained from immature xylem of loblolly pine (Pinus taeda L.). A total of 1,097 single-pass sequences were obtained from 5' ends of cDNAs made from gravistimulated tissue from bent trees. Cluster analysis detected 107 groups of similar sequences, ranging in size from 2 to 20 sequences. A total of 361 sequences fell into these groups, whereas 736 sequences were unique. About 55% of the pine EST sequences show similarity to previously described sequences in public databases. About 10% of the recognized genes encode factors involved in cell wall formation. Sequences similar to cell wall proteins, most known lignin biosynthetic enzymes, and several enzymes of carbohydrate metabolism were found. A number of putative regulatory proteins also are represented. Expression patterns of several of these genes were studied in various tissues and organs of pine. Sequencing novel genes expressed during xylem formation will provide a powerful means of identifying mechanisms controlling this important differentiation pathway.
Figures
References
-
- Dixon R K, Brown S, Houghton R A, Solomon A M, Trexler M C, Wisniewski J. Science. 1994;263:865–872. - PubMed
-
- Higuchi T. In: Biochemistry and Molecular Biology of Wood. Timell T E, editor. Heidelberg: Springer; 1997.
-
- Pigliucci M. Tree. 1996;11:168–173. - PubMed
-
- Timell T E. Compression Wood in Gymnosperms. Heidelberg: Springer; 1986.
-
- Fukushima K, Terashima N. Wood Sci Technol. 1991;25:371–381.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

