Phylogenetic analysis of methanogens from the bovine rumen
- PMID: 11384509
- PMCID: PMC32158
- DOI: 10.1186/1471-2180-1-5
Phylogenetic analysis of methanogens from the bovine rumen
Abstract
Background: Interest in methanogens from ruminants has resulted from the role of methane in global warming and from the fact that cattle typically lose 6 % of ingested energy as methane. Several species of methanogens have been isolated from ruminants. However they are difficult to culture, few have been consistently found in high numbers, and it is likely that major species of rumen methanogens are yet to be identified.
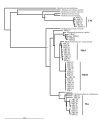
Results: Total DNA from clarified bovine rumen fluid was amplified using primers specific for Archaeal 16S rRNA gene sequences (rDNA). Phylogenetic analysis of 41 rDNA sequences identified three clusters of methanogens. The largest cluster contained two distinct subclusters with rDNA sequences similar to Methanobrevibacter ruminantium 16S rDNA. A second cluster contained sequences related to 16S rDNA from Methanosphaera stadtmanae, an organism not previously described in the rumen. The third cluster contained rDNA sequences that may form a novel group of rumen methanogens.
Conclusions: The current set of 16S rRNA hybridization probes targeting methanogenic Archaea does not cover the phylogenetic diversity present in the rumen and possibly other gastro-intestinal tract environments. New probes and quantitative PCR assays are needed to determine the distribution of the newly identified methanogen clusters in rumen microbial communities.
Figures
References
-
- Bergey DH. The methanogens. Bergey's Manual of Determinative Bacteriology Edited by Holt JG, 9 ed. pp. 719-725. Baltimore: Williams and Wilkins Co.; 1994. pp. 719–725.
-
- Johnson KA, Johnson DE. Methane emissions from cattle. J Anim Sci. 1995;73:2483–2492. - PubMed
-
- Stewart CS, Flint HJ, Bryant MP. The rumen bacteria. The Rumen Microbial Ecosystem Edited by Hobson PN, Stewart CS, 2 ed. pp. 10-72. New York: Blackie Academic and Professional; 1997. pp. 10–72.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

