Molecular cloning of an apoptosis-inducing protein, pierisin, from cabbage butterfly: possible involvement of ADP-ribosylation in its activity
- PMID: 10485873
- PMCID: PMC17930
- DOI: 10.1073/pnas.96.19.10608
Molecular cloning of an apoptosis-inducing protein, pierisin, from cabbage butterfly: possible involvement of ADP-ribosylation in its activity
Abstract
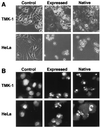
We have previously reported that the cabbage butterfly, Pieris rapae, contains a 98-kDa protein, named pierisin, that induces apoptosis in a variety of human cancer cell lines. In the present study, sequencing and cloning of a cDNA encoding pierisin was accomplished. PCR-direct sequencing showed that the gene encodes an 850-amino acid protein with a calculated molecular weight of 98,081. An intact clone at the amino acid level encompassing the entire coding region was obtained by recombination of two independent clones, and the molecular mass of its in vitro expressed protein was about 100 kDa on SDS/PAGE, the same as that of purified native pierisin. The expressed protein induced apoptosis in human gastric carcinoma TMK-1 and cervical carcinoma HeLa cells, like the native protein, indicating functional activity. The deduced amino acid sequence of pierisin showed 32% homology with a 100-kDa mosquitocidal toxin from Bacillus sphaericus SSII-1. In addition, pierisin showed regional sequence similarities with ADP-ribosylating toxins, such as the A subunit of cholera toxin. A glutamic acid residue at the putative NAD-binding site, conserved in all ADP-ribosylating toxins, was also found in pierisin. Substitution of another amino acid for glutamic acid 165 resulted in a great decrease in cytotoxicity and induction of apoptosis. Moreover, inhibitors of ADP-ribosylating enzymes reduced pierisin-induced apoptosis. These results suggest that the apoptosis-inducing protein pierisin might possess ADP-ribosylation activity that leads to apoptosis of the cells.
Figures






References
-
- Boman H G, Hultmark D. Annu Rev Microbiol. 1987;41:103–126. - PubMed
-
- Hultmark D. Trends Genet. 1993;9:178–183. - PubMed
-
- Hoffmann J A. Curr Opin Immunol. 1995;7:4–10. - PubMed
-
- Itoh A, Iizuka K, Natori S. Jpn J Cancer Res (GANN) 1985;76:1027–1033. - PubMed
-
- Moore A J, Devine D A, Bibby M C. Pept Res. 1994;7:265–269. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

