Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa
- PMID: 16326928
- PMCID: PMC1323481
- DOI: 10.1105/tpc.105.037200
Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa
Abstract
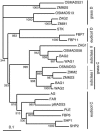
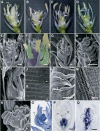
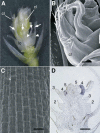
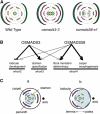
The C-class MADS box gene AGAMOUS (AG) plays crucial roles in Arabidopsis thaliana development by regulating the organ identity of stamens and carpels, the repression of A-class genes, and floral meristem determinacy. To examine the conservation and diversification of C-class gene function in monocots, we analyzed two C-class genes in rice (Oryza sativa), OSMADS3 and OSMADS58, which may have arisen by gene duplication before divergence of rice and maize (Zea mays). A knockout line of OSMADS3, in which the gene is disrupted by T-DNA insertion, shows homeotic transformation of stamens into lodicules and ectopic development of lodicules in the second whorl near the palea where lodicules do not form in the wild type but carpels develop almost normally. By contrast, RNA-silenced lines of OSMADS58 develop astonishing flowers that reiterate a set of floral organs, including lodicules, stamens, and carpel-like organs, suggesting that determinacy of the floral meristem is severely affected. These results suggest that the two C-class genes have been partially subfunctionalized during rice evolution (i.e., the functions regulated by AG have been partially partitioned into two paralogous genes, OSMADS3 and OSMADS58, which were produced by a recent gene duplication event in plant evolution).
Figures




References
-
- Alvarez-Buylla, E.R., Pelaz, S., Liljegren, S.J., Gold, S.E., Burgeff, C., Ditta, G.S., Ribas de Pouplana, L., Martinez-Castilla, L., and Yanofsky, M.F. (2000). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 97 5328–5333. - PMC - PubMed
-
- Ambrose, B.A., Lerner, D.R., Ciceri, P., Padilla, C.M., Yanofsky, M.F., and Schmidt, R.J. (2000). Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell 5 569–579. - PubMed
-
- Bommert, P., Satoh-Nagasawa, N., Jackson, D., and Hirano, H.-Y. (2005). Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol. 46 69–78. - PubMed
-
- Bowman, J.L., Baum, S.F., Eshed, Y., Putterill, J., and Alvarez, J. (1999). Molecular genetics of gynoecium development in Arabidopsis. Curr. Top. Dev. Biol. 45 155–205. - PubMed
-
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112 1–20. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

