Identification and isolation of anaerobic, syntrophic phthalate isomer-degrading microbes from methanogenic sludges treating wastewater from terephthalate manufacturing
- PMID: 15006786
- PMCID: PMC368352
- DOI: 10.1128/AEM.70.3.1617-1626.2004
Identification and isolation of anaerobic, syntrophic phthalate isomer-degrading microbes from methanogenic sludges treating wastewater from terephthalate manufacturing
Abstract
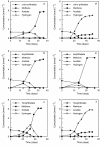
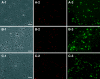
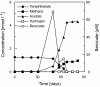
The microbial populations responsible for anaerobic degradation of phthalate isomers were investigated by enrichment and isolation of those microbes from anaerobic sludge treating wastewater from the manufacturing of terephthalic acid. Primary enrichments were made with each of three phthalate isomers (ortho-, iso-, and terephthalate) as the sole energy source at 37 degrees C with two sources of anaerobic sludge (both had been used to treat wastewater containing high concentrations of phthalate isomers) as the inoculum. Six methanogenic enrichment cultures were obtained which not only degraded the isomer used for the enrichment but also had the potential to degrade part of other phthalate isomers as well as benzoate with concomitant production of methane, presumably involving strictly syntrophic substrate degradation. Our 16S rRNA gene-cloning analysis combined with fluorescence in situ hybridization revealed that the predominant bacteria in the enrichment cultures were affiliated with a recently recognized non-sulfate-reducing subcluster (subcluster Ih) in the group 'Desulfotomaculum lineage I' or a clone cluster (group TA) in the class delta-PROTEOBACTERIA: Several attempts were made to isolate these microbes, resulting in the isolation of a terephthalate-degrading bacterium, designated strain JT, in pure culture. A coculture of the strain with the hydrogenotrophic methanogen Methanospirillum hungatei converted terephthalate to acetate and methane within 3 months of incubation, whereas strain JT could not degrade terephthalate in pure culture. During the degradation of terephthalate, a small amount of benzoate was transiently accumulated as an intermediate, indicative of decarboxylation of terephthalate to benzoate as the initial step of the degradation. 16S rRNA gene sequence analysis revealed that the strain was a member of subcluster Ih of the group 'Desulfotomaculum lineage I', but it was only distantly related to other known species.
Figures


References
-
- Alvarez, J. M., S. Macé, and P. Llabrés. 2000. Anaerobic digestion of organic solid wastes. An overview of research achievements and perspectives. Bioresource Technol. 74:3-16.
-
- Brauman, A., J. A. Müller, J. L. Garcia, A. Brune, and B. Schink. 1998. Fermentative degradation of 3-hydroxybenzoate in pure culture by a novel strictly anaerobic bacterium, Sporotomaculum hydroxybenzoicum gen. nov., sp. nov. Int. J. Syst. Bacteriol. 48:215-221. - PubMed
-
- Cord-Ruwisch, R., and J. L. Garcia. 1985. Isolation and characterization of an anaerobic benzoate-degrading spore-forming sulfate-reducing bacterium, Desulfotomaculum sapomandens sp. nov. FEMS Microbiol. Lett. 29:325-330.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

