Robust control of the seasonal expression of the Arabidopsis FLC gene in a fluctuating environment
- PMID: 20534541
- PMCID: PMC2895080
- DOI: 10.1073/pnas.0914293107
Robust control of the seasonal expression of the Arabidopsis FLC gene in a fluctuating environment
Abstract
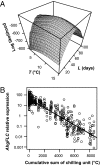
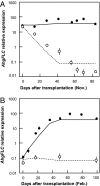
Plants flower in particular seasons even in natural, fluctuating environments. The molecular basis of temperature-dependent flowering-time regulation has been extensively studied, but little is known about how gene expression is controlled in natural environments. Without a memory of past temperatures, it would be difficult for plants to detect seasons in natural, noisy environments because temperature changes occurring within a few weeks are often inconsistent with seasonal trends. Our 2-y census of the expression of a temperature-dependent flowering-time gene, AhgFLC, in a natural population of perennial Arabidopsis halleri revealed that the regulatory system of this flowering-time gene extracts seasonal cues as if it memorizes temperatures over the past 6 wk. Time-series analysis revealed that as much as 83% of the variation in the AhgFLC expression is explained solely by the temperature for the previous 6 wk, but not by the temperatures over shorter or longer periods. The accuracy of our model in predicting the gene expression pattern under contrasting temperature regimes in the transplant experiments indicates that such modeling incorporating the molecular bases of flowering-time regulation will contribute to predicting plant responses to future climate changes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Richards CL, et al. Perspective on ecological and evolutionary systems biology. Annu Plant Rev. 2009;35:331–351.
-
- Wilczek AM, et al. Effects of genetic perturbation on seasonal life history plasticity. Science. 2009;323:930–934. - PubMed
-
- Simpson GG, Dean C. Arabidopsis, the Rosetta stone of flowering time? Science. 2002;296:285–289. - PubMed
-
- Yanovsky MJ, Kay SA. Living by the calendar: How plants know when to flower. Nat Rev Mol Cell Biol. 2003;4:265–275. - PubMed
-
- He YH, Amasino RM. Role of chromatin modification in flowering-time control. Trends Plant Sci. 2005;10:30–35. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

