Horizontal chromosome transfer, a mechanism for the evolution and differentiation of a plant-pathogenic fungus
- PMID: 19749175
- PMCID: PMC2772402
- DOI: 10.1128/EC.00135-09
Horizontal chromosome transfer, a mechanism for the evolution and differentiation of a plant-pathogenic fungus
Abstract
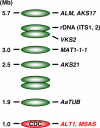
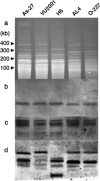
The tomato pathotype of Alternaria alternata produces host-specific AAL toxin and causes Alternaria stem canker on tomato. A polyketide synthetase (PKS) gene, ALT1, which is involved in AAL toxin biosynthesis, resides on a 1.0-Mb conditionally dispensable chromosome (CDC) found only in the pathogenic and AAL toxin-producing strains. Genomic sequences of ALT1 and another PKS gene, both of which reside on the CDC in the tomato pathotype strains, were compared to those of tomato pathotype strains collected worldwide. This revealed that the sequences of both CDC genes were identical among five A. alternata tomato pathotype strains having different geographical origins. On the other hand, the sequences of other genes located on chromosomes other than the CDC are not identical in each strain, indicating that the origin of the CDC might be different from that of other chromosomes in the tomato pathotype. Telomere fingerprinting and restriction fragment length polymorphism analyses of the A. alternata strains also indicated that the CDCs in the tomato pathotype strains were identical, although the genetic backgrounds of the strains differed. A hybrid strain between two different pathotypes was shown to harbor the CDCs derived from both parental strains with an expanded range of pathogenicity, indicating that CDCs can be transmitted from one strain to another and stably maintained in the new genome. We propose a hypothesis whereby the ability to produce AAL toxin and to infect a plant could potentially be distributed among A. alternata strains by horizontal transfer of an entire pathogenicity chromosome. This could provide a possible mechanism by which new pathogens arise in nature.
Figures






References
-
- Adachi, Y., and T. Tsuge. 1994. Coinfection by different isolates of Alternaria alternata in single black spot lesions of Japanese pear leaves. Phytopathology 84:447-451.
-
- Akagi, Y., M. Taga, M. Yamamoto, T. Tsuge, Y. Fukumasa-Nakai, H. Otani, and M. Kodama. 2009. Chromosome constitution of hybrid strains constructed by protoplast fusion between the tomato and strawberry pathotypes of Alternaria alternata. J. Gen. Plant Pathol. 75:101-109.
-
- Akamatsu, H., Y. Itoh, M. Kodama, H. Otani, and K. Kohmoto. 1997. AAL-toxin deficient mutants of Alternaria alternata tomato pathotype by restriction enzyme-mediated integration. Phytopathology 87:967-972. - PubMed
-
- Akamatsu, H., H. Otani, and M. Kodama. 2003. Characterization of a gene cluster for host-specific AAL-toxin biosynthesis in the tomato pathotype of Alternaria alternata. Fungal Genet. Newsl. 50(Suppl.):355.
-
- Akamatsu, H., M. Taga, M. Kodama, R. Johnson, H. Otani, and K. Kohmoto. 1999. Molecular karyotypes for Alternaria plant pathogens known to produce host-specific toxins. Curr. Genet. 35:647-656. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

