Extensive duplication and reshuffling in the Arabidopsis genome
- PMID: 10899976
- PMCID: PMC149051
- DOI: 10.1105/tpc.12.7.1093
Extensive duplication and reshuffling in the Arabidopsis genome
Abstract
Systematic analysis of the Arabidopsis genome provides a basis for detailed studies of genome structure and evolution. Members of multigene families were mapped, and random sequence alignment was used to identify regions of extended similarity in the Arabidopsis genome. Detailed analysis showed that the number, order, and orientation of genes were conserved over large regions of the genome, revealing extensive duplication covering the majority of the known genomic sequence. Fine mapping analysis showed much rearrangement, resulting in a patchwork of duplicated regions that indicated deletion, insertion, tandem duplication, inversion, and reciprocal translocation. The implications of these observations for evolution of the Arabidopsis genome as well as their usefulness for analysis and annotation of the genomic sequence and in comparative genomics are discussed.
Figures


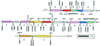
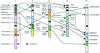
References
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. - PubMed
-
- Ansari-Lari, M.A., Oeltjen, J.C., Schwartz, S., Zhang, Z., Muzny, D.M., Lu, J., Gorrell, J.H., Chinault, A.C., Belmont, J.W., Miller, W., and Gibbs, R.A. (1998). Comparative sequence analysis of a gene-rich cluster at human chromosome 12p13 and its syntenic region in mouse chromosome 6. Genome Res. 8, 29–40. - PubMed
-
- Axelos, M., Bardet, C., Liboz, T., Le Van Thai, A., Curie, C., and Lescure, B. (1989). The gene family encoding the Arabidopsis thaliana translation elongation factor EF-1α: Molecular cloning, characterization and expression. Mol. Gen. Genet. 219, 106–112. - PubMed
-
- Bevan, M., et al. (1998). Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 391, 485–488. - PubMed
-
- Cavell, A.C., Lydiate, D.J., Parkin, I.A., Dean, C., and Trick, M. (1998). Collinearity between a 30-centimorgan segment of Arabidopsis thaliana chromosome 4 and duplicated regions within the Brassica napus genome. Genome 41, 62–69. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

