The breakpoint region of the most common isochromosome, i(17q), in human neoplasia is characterized by a complex genomic architecture with large, palindromic, low-copy repeats
- PMID: 14666446
- PMCID: PMC1181896
- DOI: 10.1086/380648
The breakpoint region of the most common isochromosome, i(17q), in human neoplasia is characterized by a complex genomic architecture with large, palindromic, low-copy repeats
Abstract
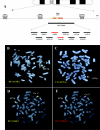
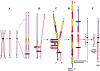
Although a great deal of information has accumulated regarding the mechanisms underlying constitutional DNA rearrangements associated with inherited disorders, virtually nothing is known about the molecular processes involved in acquired neoplasia-associated chromosomal rearrangements. Isochromosome 17q, or "i(17q)," is one of the most common structural abnormalities observed in human neoplasms. We previously identified a breakpoint cluster region for i(17q) formation in 17p11.2 and hypothesized that genome architectural features could be responsible for this clustering. To address this hypothesis, we precisely mapped the i(17q) breakpoints in 11 patients with different hematologic malignancies and determined the genomic structure of the involved region. Our results reveal a complex genomic architecture in the i(17q) breakpoint cluster region, characterized by large ( approximately 38-49-kb), palindromic, low-copy repeats, strongly suggesting that somatic rearrangements are not random events but rather reflect susceptibilities due to the genomic structure.
Figures

References
Electronic-Database Information
-
- Blast 2 Sequences, http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html (for the Blast 2 browser)
-
- GenBank, http://www.ncbi.nih.gov/Genbank/ (for CTC-457L16 [accession number AC003957], RP11-160E2 [accession number AC007952], RP11-744A16 [accession number AC106017], RP11-970O14 [accession number AC036110], CTB-187M2 [accession number AC004448], RP11-1113L8 [accession number AC025627], CTD-2354J3 [accession number AC015935], RP11-311F12 [accession number AC005722], RP11-78O7 [accession number AC015726], RP11-209D14 [accession number AC005730], CTD-2010G8 [accession number AC007963], RP1-149D14 [accession number AJ009617], CTD-2525F10 [accession number AC109313], RP11-135L13 [accession number AC124066], L29232 [accession number AC139083], L29246 [accession number AC139093], L29227 [accession number AC139315], L29233 [accession number AC140106], L29228 [accession number AC144507], L29248 [accession number AC139138], L29225 [accession number AC144506], L29280 [accession number AC139077], mouse RP23-278F12 [accession number AC084044], rat CH230-200D5 [accession number AC105503], and rat CH230-255K6 [accession number AC134746])
-
- Mitelman Database of Chromosome Aberrations in Cancer, http://cgap.nci.nih.gov/Chromosomes/Mitelman
-
- RepeatMasker Web Server, http://repeatmasker.genome.washington.edu/cgi-bin/RepeatMasker
References
-
- Barbouti A, Johansson B, Höglund M, Mauritzson N, Strömbeck B, Nilsson PG, Tanke HJ, Hagemeijer A, Mitelman F, Fioretos T (2002) Multicolor COBRA-FISH analysis of chronic myeloid leukemia reveals novel cryptic balanced translocations during disease progression. Genes Chromosomes Cancer 35:127–13710.1002/gcc.10099 - DOI - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

