Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa
- PMID: 16632644
- PMCID: PMC1475497
- DOI: 10.1105/tpc.105.040535
Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa
Abstract
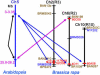
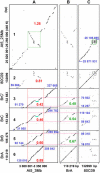
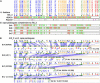
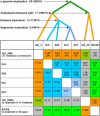
Strong evidence exists for polyploidy having occurred during the evolution of the tribe Brassiceae. We show evidence for the dynamic and ongoing diploidization process by comparative analysis of the sequences of four paralogous Brassica rapa BAC clones and the homologous 124-kb segment of Arabidopsis thaliana chromosome 5. We estimated the times since divergence of the paralogous and homologous lineages. The three paralogous subgenomes of B. rapa triplicated 13 to 17 million years ago (MYA), very soon after the Arabidopsis and Brassica divergence occurred at 17 to 18 MYA. In addition, a pair of BACs represents a more recent segmental duplication, which occurred approximately 0.8 MYA, and provides an exception to the general expectation of three paralogous segments within the B. rapa genome. The Brassica genome segments show extensive interspersed gene loss relative to the inferred structure of the ancestral genome, whereas the Arabidopsis genome segment appears little changed. Representatives of all 32 genes in the Arabidopsis genome segment are represented in Brassica, but the hexaploid complement of 96 has been reduced to 54 in the three subgenomes, with compression of the genomic region lengths they occupy to between 52 and 110 kb. The gene content of the recently duplicated B. rapa genome segments is identical, but intergenic sequences differ.
Figures

References
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. - PubMed
-
- Babula, D., Kaczmarek, M., Barakat, A., Delseny, M., Quiros, C.F., and Sadowski, J. (2003). Chromosomal mapping of Brassica oleracea based on ESTs from Arabidopsis thaliana: Complexity of the comparative map. Mol. Genet. Genomics 268 656–665. - PubMed
-
- Bennetzen, J.L. (2000). Transposable element contributions to plant gene and genome evolution. Plant Mol. Biol. 42 251–269. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

