Genetic divergence of the rhesus macaque major histocompatibility complex
- PMID: 15289473
- PMCID: PMC509259
- DOI: 10.1101/gr.2134504
Genetic divergence of the rhesus macaque major histocompatibility complex
Abstract
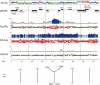
The major histocompatibility complex (MHC) is comprised of the class I, class II, and class III regions, including the MHC class I and class II genes that play a primary role in the immune response and serve as an important model in studies of primate evolution. Although nonhuman primates contribute significantly to comparative human studies, relatively little is known about the genetic diversity and genomics underlying nonhuman primate immunity. To address this issue, we sequenced a complete rhesus macaque MHC spanning over 5.3 Mb, and obtained an additional 2.3 Mb from a second haplotype, including class II and portions of class I and class III. A major expansion of from six class I genes in humans to as many as 22 active MHC class I genes in rhesus and levels of sequence divergence some 10-fold higher than a similar human comparison were found, averaging from 2% to 6% throughout extended portions of class I and class II. These data pose new interpretations of the evolutionary constraints operating between MHC diversity and T-cell selection by contrasting with models predicting an optimal number of antigen presenting genes. For the clinical model, these data and derivative genetic tools can be implemented in ongoing genetic and disease studies that involve the rhesus macaque.
Copyright 2004 Cold Spring Harbor Laboratory Press ISSN
Figures



References
-
- Accolla, R.S., Adorini, L., Sartoris, S., Sinigaglia, F., and Guardiola, J. 1995. MHC: Orchestrating the immune response. Immunol. Today 16: 8–11. - PubMed
-
- Adams, E.J. and Parham, P. 2001. Species-specific evolution of MHC class I genes in the higher primates. Immunol. Rev. 183: 41–64. - PubMed
-
- Anzai, T., Shiina, T., Kimura, N., Yanagiya, K., Kohara, S., Shigenari, A., Yamagata, T., Kulski, J.K., Naruse, T.K., Fujimori, Y., et al. 2003. Comparative sequencing of human and chimpanzee MHC class I regions unveils insertions/deletions as the major path to genomic divergence. Proc. Natl. Acad. Sci. 100: 7708–7713. - PMC - PubMed
-
- Austad, S.N. 1997. Small nonhuman primates as potential models of human aging. Ilar J. 38: 142–147. - PubMed
-
- Bontrop, R.E. 2001. Non-human primates: Essential partners in biomedical research. Immunol. Rev. 183: 5–9. - PubMed
WEB SITE REFERENCES
-
- http://www.sanger.ac.uk/HGP/Chr6/MHC/; The Sanger Institute/The MHC haplotype project.
-
- http://deepc2.zool.iastate.edu/aat/map/mapdoc.html; Information resource on the Multiple Sequence Alignment.
-
- www.fhcrc.org/labs/geraghty; Geraghty lab Web data resource.
-
- www.chori.org/bacpac/; BACPAC Resources center homepage.
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
