Rapid reorganization of resistance gene homologues in cereal genomes
- PMID: 9419382
- PMCID: PMC18227
- DOI: 10.1073/pnas.95.1.370
Rapid reorganization of resistance gene homologues in cereal genomes
Abstract
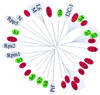
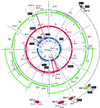
We used conserved domains in the major class (nucleotide binding site plus leucine-rich repeat) of dicot resistance (R) genes to isolate related gene fragments via PCR from the monocot species rice and barley. Peptide sequence comparison of dicot R genes and monocot R-like genes revealed shared motifs but provided no evidence for a monocot-specific signature. Mapping of these genes in rice and barley showed linkage to genetically characterized R genes and revealed the existence of mixed clusters, each harboring at least two highly dissimilar R-like genes. Diversity was detected intraspecifically with wide variation in copy number between varieties of a particular species. Interspecific analyses of R-like genes frequently revealed nonsyntenic map locations between the cereal species rice, barley, and foxtail millet although tight collinear gene order is a hallmark of monocot genomes. Our data suggest a dramatic rearrangement of R gene loci between related species and implies a different mechanism for nucleotide binding site plus leucine-rich repeat gene evolution compared with the rest of the monocot genome.
Figures

References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

