Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution
- PMID: 9495754
- PMCID: PMC107003
- DOI: 10.1128/JB.180.5.1159-1165.1998
Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution
Abstract
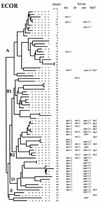
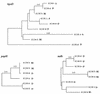
We studied the ancestry of virulence-associated genes in Escherichia coli by examining chromosomal regions specific to pathogenic isolates. The four virulence determinants examined were the alpha-hemolysin (hly) loci hlyI and hlyII, the type II capsule gene cluster kps, and the P (pap) and S (sfa) fimbria gene clusters. All four loci were shown previously to be associated with pathogenicity islands of uropathogenic E. coli isolates. The hly, kps, sfa, and pap regions each have an unexpected clustered distribution among the E. coli collection of reference (ECOR) strains, but all these regions were absent from a collection of diarrheagenic E. coli isolates. Strains in the ECOR subgroup B2 typically had a combination of at least three of the four loci, and all strains in subgroup D had a copy of the kps and pap clusters. In contrast, only four strains in subgroup A had either hly, kps, sfa, or pap, and no subgroup A strains had all four together. Strains of subgroup B1 were devoid of all four virulence regions, with the exception of one isolate that had a copy of the sfa gene cluster. This phylogenetic distribution of strain-specific sequences corresponds to the ECOR groups with the largest genome size, namely, B2 and D. We propose that the pathogenicity islands are ancestral to subgroups B2 and D and were acquired after speciation, with subsequent horizontal transfer into some group A, B1, and E lineages. These results suggest that the hly, kps, sfa, and pap pathogenicity determinants may play a role in the evolution of enteric bacteria quite apart from, and perhaps with precedence over, their ability to cause disease.
Figures


References
-
- Blum G, Falbo V, Caprioli A, Hacker J. Gene clusters encoding the cytotoxic necrotizing factor type 1, Prs-fimbriae and alpha-hemolysin form the pathogenicity island II of the uropathogenic Escherichia coli strain J96. FEMS Microbiol Lett. 1995;126:189–195. - PubMed
-
- Boulnois G J, Roberts I S. Genetics of capsular polysaccharide production in bacteria. Curr Top Microbiol Immunol. 1990;150:1–18. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

