Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation
- PMID: 9758812
- PMCID: PMC106571
- DOI: 10.1128/AEM.64.10.3869-3877.1998
Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation
Abstract
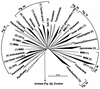
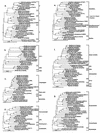
A culture-independent molecular phylogenetic approach was used to survey constituents of microbial communities associated with an aquifer contaminated with hydrocarbons (mainly jet fuel) and chlorinated solvents undergoing intrinsic bioremediation. Samples were obtained from three redox zones: methanogenic, methanogenic-sulfate reducing, and iron or sulfate reducing. Small-subunit rRNA genes were amplified directly from aquifer material DNA by PCR with universally conserved or Bacteria- or Archaea-specific primers and were cloned. A total of 812 clones were screened by restriction fragment length polymorphisms (RFLP), approximately 50% of which were unique. All RFLP types that occurred more than once in the libraries, as well as many of the unique types, were sequenced. A total of 104 (94 bacterial and 10 archaeal) sequence types were determined. Of the 94 bacterial sequence types, 10 have no phylogenetic association with known taxonomic divisions and are phylogenetically grouped in six novel division level groups (candidate divisions WS1 to WS6); 21 belong to four recently described candidate divisions with no cultivated representatives (OP5, OP8, OP10, and OP11); and 63 are phylogenetically associated with 10 well-recognized divisions. The physiology of two particularly abundant sequence types obtained from the methanogenic zone could be inferred from their phylogenetic association with groups of microorganisms with a consistent phenotype. One of these sequence types is associated with the genus Syntrophus; Syntrophus spp. produce energy from the anaerobic oxidation of organic acids, with the production of acetate and hydrogen. The organism represented by the other sequence type is closely related to Methanosaeta spp., which are known to be capable of energy generation only through aceticlastic methanogenesis. We hypothesize, therefore, that the terminal step of hydrocarbon degradation in the methanogenic zone of the aquifer is aceticlastic methanogenesis and that the microorganisms represented by these two sequence types occur in syntrophic association.
Figures


References
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Barcelona M J, Lu J, Tomczak D M. Organic acid derivatization techniques applied to petroleum hydrocarbon transformation in surface environments. Ground Water Monit Remed. 1995;15:114–124.
-
- Borden R C. Natural bioremediation of hydrocarbon-contaminated ground water. In: Norris R D, et al., editors. Handbook of bioremediation. Boca Raton, Fla: CRC Press, Inc.; 1994. pp. 177–199.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

