Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization
- PMID: 9765550
- PMCID: PMC34840
- DOI: 10.1104/pp.118.2.637
Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization
Abstract
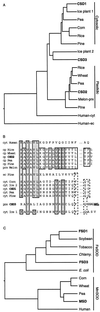
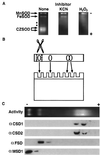
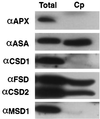
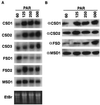
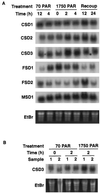
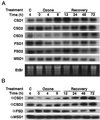
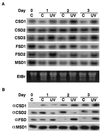
A number of environmental stresses can lead to enhanced production of superoxide within plant tissues, and plants are believed to rely on the enzyme superoxide dismutase (SOD) to detoxify this reactive oxygen species. We have identified seven cDNAs and genes for SOD in Arabidopsis. These consist of three CuZnSODs (CSD1, CSD2, and CSD3), three FeSODs (FSD1, FSD2, and FSD3), and one MnSOD (MSD1). The chromosomal location of these seven SOD genes has been established. To study this enzyme family, antibodies were generated against five proteins: CSD1, CSD2, CSD3, FSD1, and MSD1. Using these antisera and nondenaturing-polyacrylamide gel electrophoresis enzyme assays, we identified protein and activity for two CuZnSODs and for FeSOD and MnSOD in Arabidopsis rosette tissue. Additionally, subcellular fractionation studies revealed the presence of CSD2 and FeSOD protein within Arabidopsis chloroplasts. The seven SOD mRNAs and the four proteins identified were differentially regulated in response to various light regimes, ozone fumigation, and ultraviolet-B irradiation. To our knowledge, this is the first report of a large-scale analysis of the regulation of multiple SOD proteins in a plant species.
Figures
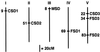



References
-
- Alscher RG, Donahue JL, Cramer CL. Reactive oxygen species and antioxidants: relationships in green cells. Physiol Plant. 1997;100:224–233.
-
- Alscher RG, Hess JL. Antioxidants in Higher Plants. Boca Raton, FL: CRC Press; 1993.
-
- Babior BM, El Benna J, Chanock SJ, Smith RM. The NADPH oxidase of leukocytes: The respiratory burst oxidase. In: Scandalios JG, editor. Oxidative Stress and the Molecular Biology of Antioxidant Defenses, Vol 34. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 737–783.
-
- Beauchamp C, Fridovich I. Superoxide dismutase: improved assays and assay applicable to acrylamide gels. Anal Biochem. 1971;44:276–287. - PubMed
-
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

