The pspC gene of Streptococcus pneumoniae encodes a polymorphic protein, PspC, which elicits cross-reactive antibodies to PspA and provides immunity to pneumococcal bacteremia
- PMID: 10569772
- PMCID: PMC97064
- DOI: 10.1128/IAI.67.12.6533-6542.1999
The pspC gene of Streptococcus pneumoniae encodes a polymorphic protein, PspC, which elicits cross-reactive antibodies to PspA and provides immunity to pneumococcal bacteremia
Abstract
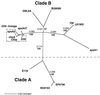
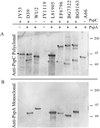
PspC is one of three designations for a pneumococcal surface protein whose gene is present in approximately 75% of all Streptococcus pneumoniae strains. Under the name SpsA, the protein has been shown to bind secretory immunoglobulin A (S. Hammerschmidt, S. R. Talay, P. Brandtzaeg, and G. S. Chhatwal, Mol. Microbiol. 25:1113-1124, 1997). Under the name CbpA, the protein has been shown to interact with human epithelial and endothelial cells (C. Rosenow et al., Mol. Microbiol. 25:819-829, 1997). The gene is paralogous to the pspA gene in S. pneumoniae and was thus called pspC (A. Brooks-Walter, R. C. Tart, D. E. Briles, and S. K. Hollingshead, Abstracts of the 97th General Meeting of the American Society for Microbiology 1997). Sequence comparisons of five published and seven new alleles reveal that this gene has a mosaic structure, and modular domains have contributed to gene diversity during evolution. Two major clades exist: clade A alleles are larger and contain an extra module that is shared with many pspA alleles; clade B alleles are smaller and lack this pspA-like domain. All alleles have a proline-rich domain and a choline-binding repeat domain that show 0% divergence from similar domains in the PspA protein. Immunization of a rabbit with a recombinant clade B PspC molecule produced antiserum that cross-reacted with both PspC and PspA from 15 pneumococcal isolates. The cross-reactive antibodies afforded cross-protection in a mouse model system. Mice immunized with PspC were protected against challenge with a strain that expressed PspA but not PspC. The PspA- and PspC-cross-reactive antibodies were directed to the proline-rich domain present in both molecules.
Figures








References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1987.
-
- Breise T, Hackenbeck R. Interactions of pneumococcal amidase with lipoteichoic acid and choline. Eur J Biochem. 1985;146:417–427. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

