Investigation of the Streptomyces clavuligerus cephamycin C gene cluster and its regulation by the CcaR protein
- PMID: 9696752
- PMCID: PMC107400
- DOI: 10.1128/JB.180.16.4068-4079.1998
Investigation of the Streptomyces clavuligerus cephamycin C gene cluster and its regulation by the CcaR protein
Abstract
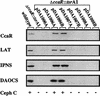
As part of a search for transcriptional regulatory genes, sequence analysis of several previously unsequenced gaps in the cephamycin biosynthetic cluster has revealed the presence in Streptomyces clavuligerus of seven genes not previously described. These include genes encoding an apparent penicillin binding protein and a transport or efflux protein, as well as the CmcI and CmcJ proteins, which catalyze late reactions in the cephamycin biosynthetic pathway. In addition, we discovered a gene, designated pcd, which displays significant homology to genes encoding semialdehyde dehydrogenases and may represent the gene encoding the long-sought-after dehydrogenase involved in the conversion of lysine to alpha-aminoadipate. Finally, two genes, sclU and rhsA, with no obvious function in cephamycin biosynthesis may define the end of the cluster. The previously described CcaR protein displays homology to a number of Streptomyces pathway-specific transcriptional activators. The ccaR gene was shown to be essential for the biosynthesis of cephamycin, clavulanic acid, and non-clavulanic acid clavams. Complementation of a deletion mutant lacking ccaR and the adjacent orf11 and blp genes showed that only ccaR was essential for the biosynthesis of cephamycin, clavulanic acid, and clavams and that mutations in orf11 or blp had no discernible effects. The lack of cephamycin production in ccaR mutants was directly attributable to the absence of biosynthetic enzymes responsible for the early and middle steps of the cephamycin biosynthetic pathway. Complementation of the ccaR deletion mutant resulted in the return of these biosynthetic enzymes and the restoration of cephamycin production.
Figures




References
-
- Aidoo K A, Wong A, Alexander D C, Rittammer R A R, Jensen S E. Cloning, sequencing and disruption of a gene from Streptomyces clavuligerus involved in clavulanic acid biosynthesis. Gene. 1994;147:41–46. - PubMed
-
- Aidoo, K. A., and S. E. Jensen. Unpublished data.
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

