A homolog of CcpA mediates catabolite control in Listeria monocytogenes but not carbon source regulation of virulence genes
- PMID: 9829942
- PMCID: PMC107718
- DOI: 10.1128/JB.180.23.6316-6324.1998
A homolog of CcpA mediates catabolite control in Listeria monocytogenes but not carbon source regulation of virulence genes
Abstract
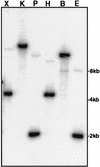
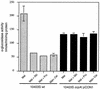
Readily utilizable sugars down-regulate virulence gene expression in Listeria monocytogenes, which has led to the proposal that this regulation may be an aspect of global catabolite regulation (CR). We recently demonstrated that the metabolic enzyme alpha-glucosidase is under CR in L. monocytogenes. Here, we report the cloning and characterization from L. monocytogenes of an apparent ortholog of ccpA, which encodes an important mediator of CR in several low-G+C-content gram-positive bacteria. L. monocytogenes ccpA (ccpALm) is predicted to encode a 335-amino-acid protein with nearly 65% identity to the gene product of Bacillus subtilis ccpA (ccpABs). Southern blot analysis with a probe derived from ccpALm revealed a single strongly hybridizing band and also a second band of much lower intensity, suggesting that there may be other closely related sequences in the L. monocytogenes chromosome, as is the case in B. subtilis. Disruption of ccpALm resulted in the inability of the mutant to grow on glucose-containing minimal medium or increase its growth rate in the presence of preferred sugars, and it completely eliminated CR of alpha-glucosidase activity in liquid medium. However, alpha-glucosidase activity was only partially relieved from CR on solid medium. These results suggest that ccpA is an important element of carbon source regulation in L. monocytogenes. Nevertheless, utilizable sugars still down-regulate the expression of hly, which encodes the virulence factor hemolysin, in a ccpALm mutant, indicating that CcpA is not involved in carbon source regulation of virulence genes.
Figures




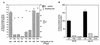
References
-
- Barak I, Behari J, Olmedo G, Guzman P, Brown D P, Castro E, Walker D, Westpheling J, Youngman P. Structure and function of the Bacillus SpoIIE protein and its localization to sites of sporulation septum assembly. Mol Microbiol. 1996;19:1047–1060. - PubMed
-
- Behari, J., and P. Youngman. Unpublished data.
-
- Bockmann R, Dickneite C, Middendorf B, Goebel W, Sokolovic Z. Specific binding of the Listeria monocytogenes transcriptional regulator PrfA to target sequences requires additional factor(s) and is influenced by iron. Mol Microbiol. 1996;22:643–653. - PubMed
-
- Bohne J, Kestler H, Uebele C, Sokolovic Z, Goebel W. Differential regulation of the virulence genes of Listeria monocytogenes by the transcriptional activator PrfA. Mol Microbiol. 1996;20:1189–1198. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

