Saprophytic and pathogenic fungi in the Ceratocystidaceae differ in their ability to metabolize plant-derived sucrose
- PMID: 26643441
- PMCID: PMC4672557
- DOI: 10.1186/s12862-015-0550-7
Saprophytic and pathogenic fungi in the Ceratocystidaceae differ in their ability to metabolize plant-derived sucrose
Abstract
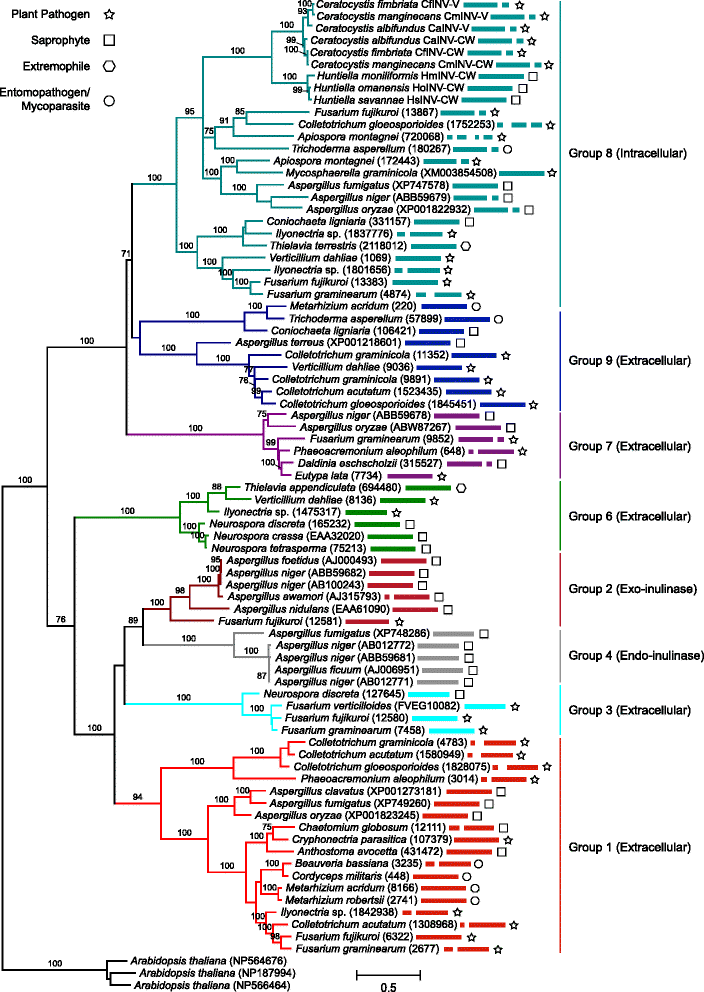
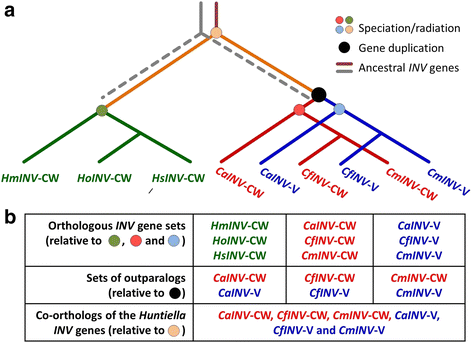
Background: Proteins in the Glycoside Hydrolase family 32 (GH32) are carbohydrate-active enzymes known as invertases that hydrolyse the glycosidic bonds of complex saccharides. Fungi rely on these enzymes to gain access to and utilize plant-derived sucrose. In fungi, GH32 invertase genes are found in higher copy numbers in the genomes of pathogens when compared to closely related saprophytes, suggesting an association between invertases and ecological strategy. The aim of this study was to investigate the distribution and evolution of GH32 invertases in the Ceratocystidaceae using a comparative genomics approach. This fungal family provides an interesting model to study the evolution of these genes, because it includes economically important pathogenic species such as Ceratocystis fimbriata, C. manginecans and C. albifundus, as well as saprophytic species such as Huntiella moniliformis, H. omanensis and H. savannae.
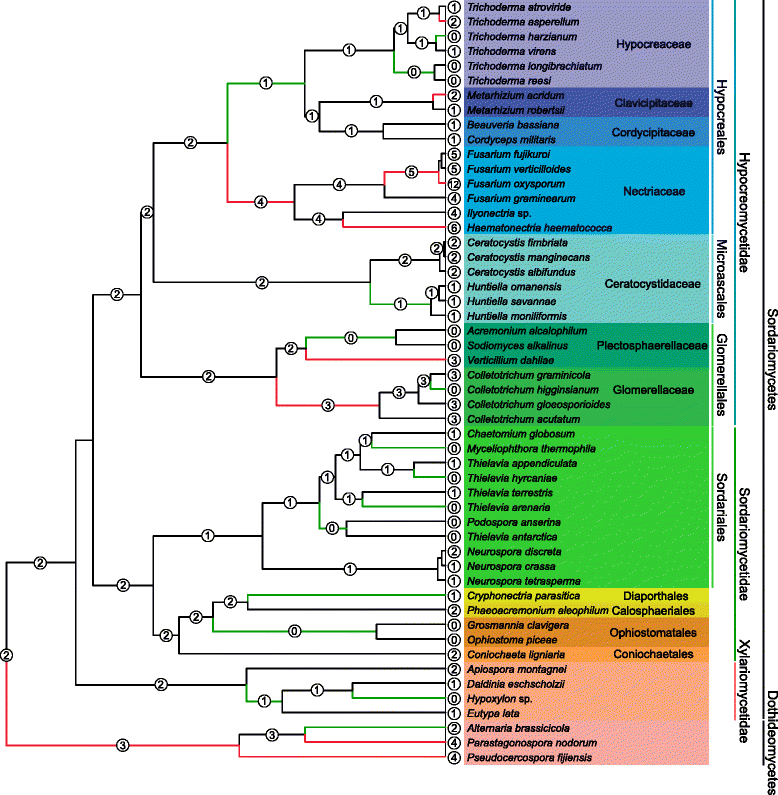
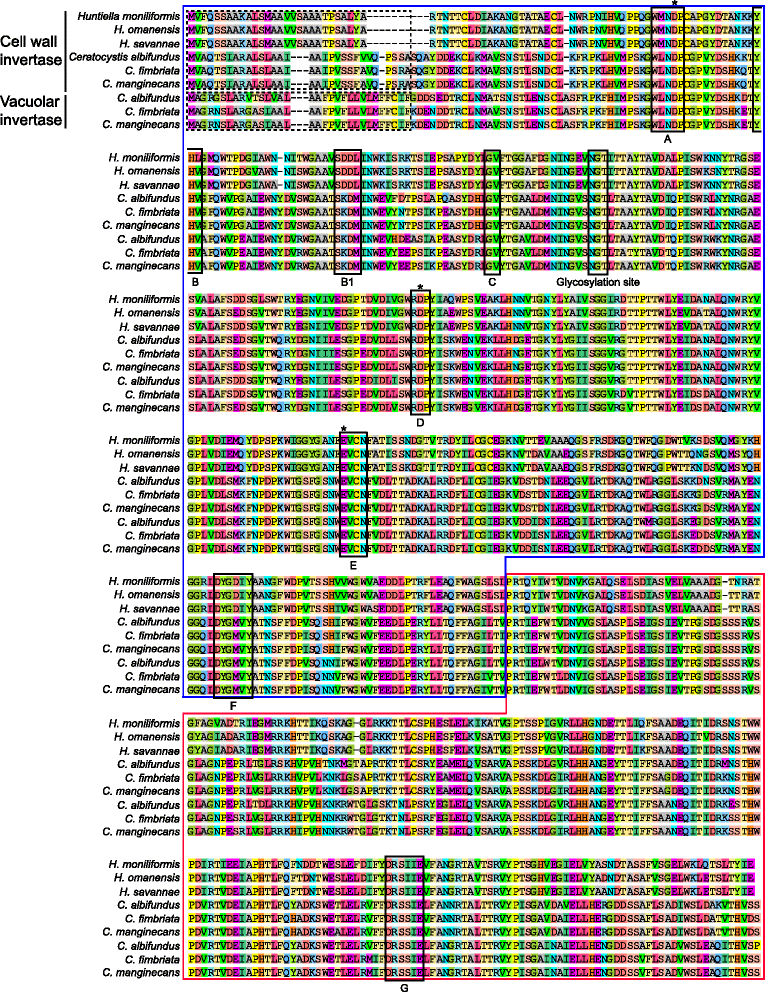
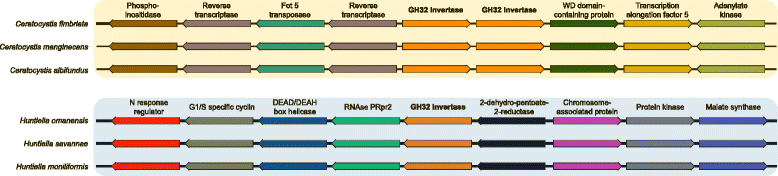
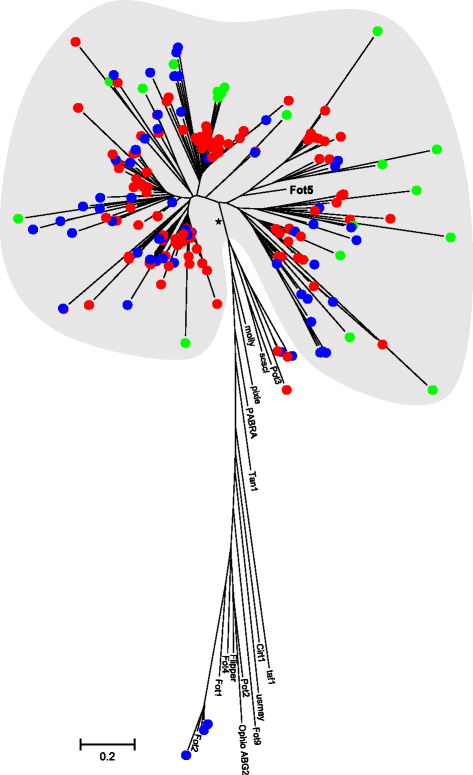
Results: The publicly available Ceratocystidaceae genome sequences, as well as the H. savannae genome sequenced here, allowed for the identification of novel GH32-like sequences. The de novo assembly of the H. savannae draft genome consisted of 28.54 megabases that coded for 7 687 putative genes of which one represented a GH32 family member. The number of GH32 gene family members appeared to be related to the ecological adaptations of these fungi. The pathogenic Ceratocystis species all contained two GH32 family genes (a putative cell wall and a putative vacuolar invertase), while the saprophytic Huntiella species had only one of these genes (a putative cell wall invertase). Further analysis showed that the evolution of the GH32 gene family in the Ceratocystidaceae involved transposable element-based retro-transposition and translocation. As an example, the activity of a Fot5-like element likely facilitated the assembly of the genomic regions harbouring the GH32 family genes in Ceratocystis.
Conclusions: This study provides insight into the evolutionary history of the GH32 gene family in Ceratocystidaceae. Our findings suggest that transposable elements shaped the evolution of the GH32 gene family, which in turn determines the sucrolytic activities and related ecological strategies of the Ceratocystidaceae species that harbour them. The study also provides insights into the role of carbohydrate-active enzymes in plant-fungal interactions and adds to our understanding of the evolution of these enzymes and their role in the life style of these fungi.
Figures

References
-
- Alberto F, Bignon C, Sulzenbacher G, Henrissat B, Czjzek M. The three-dimensional structure of invertase (β-fructosidase) from Thermotoga maritima reveals a bimodular arrangement and an evolutionary relationship between retaining and inverting glycosidases. J Biol Chem. 2004;279(18):18903–18910. doi: 10.1074/jbc.M313911200. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

